
Rhizobium sp. Leaf371
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
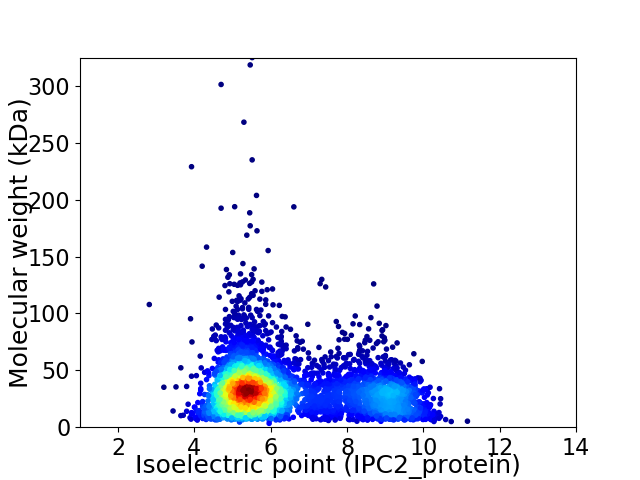
Virtual 2D-PAGE plot for 4411 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
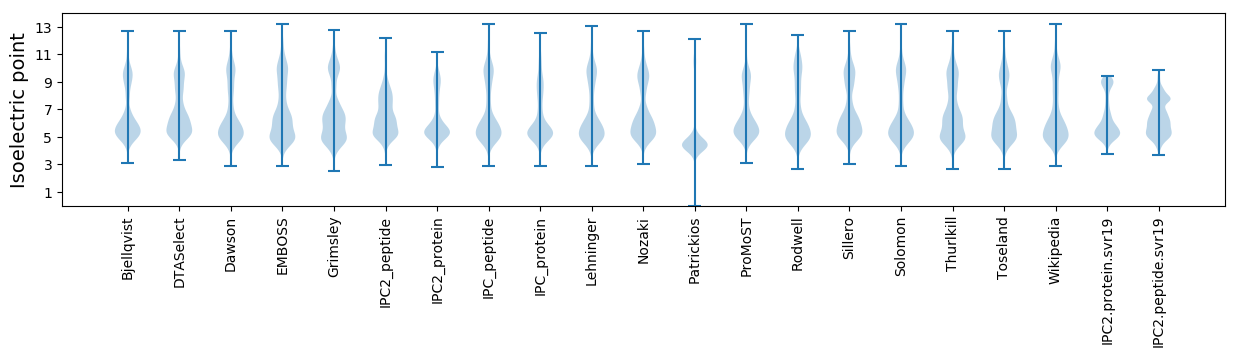
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5WSJ8|A0A0Q5WSJ8_9RHIZ ABC transporter ATP-binding protein OS=Rhizobium sp. Leaf371 OX=1736355 GN=ASG39_20835 PE=4 SV=1
MM1 pKa = 7.5IANTGNPLIDD11 pKa = 4.14GLLSGNAWNAATLTYY26 pKa = 10.66AFPTTTSAYY35 pKa = 8.41TYY37 pKa = 10.29TSGTYY42 pKa = 9.77RR43 pKa = 11.84DD44 pKa = 3.78VAPVSLAQQKK54 pKa = 10.11AALFFLEE61 pKa = 4.45QSSGPAANDD70 pKa = 3.56GFSVEE75 pKa = 4.96GFTLLKK81 pKa = 10.06FAAGSATNATLRR93 pKa = 11.84FAEE96 pKa = 4.93SPTADD101 pKa = 3.23PTAYY105 pKa = 10.37AFYY108 pKa = 10.14PGNYY112 pKa = 9.49DD113 pKa = 3.06SAGDD117 pKa = 3.57VWFGTAYY124 pKa = 10.52AGTEE128 pKa = 3.65YY129 pKa = 10.51DD130 pKa = 3.62YY131 pKa = 11.18RR132 pKa = 11.84SPKK135 pKa = 9.36IGTYY139 pKa = 9.13AWHH142 pKa = 6.27TMGHH146 pKa = 6.52EE147 pKa = 4.12IGHH150 pKa = 6.44ALGLKK155 pKa = 9.81HH156 pKa = 5.8GHH158 pKa = 6.35EE159 pKa = 4.67DD160 pKa = 3.13AALPSNVDD168 pKa = 3.4SVEE171 pKa = 3.88YY172 pKa = 10.86SIMTYY177 pKa = 9.9RR178 pKa = 11.84AYY180 pKa = 9.74IGAPLTGYY188 pKa = 10.65SYY190 pKa = 11.62GLNDD194 pKa = 3.76APQSFMSLDD203 pKa = 3.34IAALQAMYY211 pKa = 10.4GADD214 pKa = 3.49YY215 pKa = 10.65TVMNGNTVYY224 pKa = 10.67RR225 pKa = 11.84WTPDD229 pKa = 2.78SGQTIIDD236 pKa = 3.84GVVTIAPGANRR247 pKa = 11.84IFATIWDD254 pKa = 4.11GGGTDD259 pKa = 3.77TFDD262 pKa = 3.19LSAYY266 pKa = 7.55KK267 pKa = 9.52TALSIDD273 pKa = 4.13LQPGKK278 pKa = 10.88YY279 pKa = 9.88SLFSTGQTADD289 pKa = 3.64LGGGPNNGHH298 pKa = 6.24ARR300 pKa = 11.84GNIFNALLYY309 pKa = 10.7KK310 pKa = 10.9NNTASLIEE318 pKa = 4.04NVLGGSGNDD327 pKa = 3.14RR328 pKa = 11.84IVGNQVQNKK337 pKa = 9.57LFGNAGNDD345 pKa = 3.33KK346 pKa = 11.0LFGLTGNDD354 pKa = 2.97ILVGGIGADD363 pKa = 3.53TLDD366 pKa = 4.42GGSGTDD372 pKa = 3.1TASYY376 pKa = 10.32EE377 pKa = 4.03DD378 pKa = 3.78AKK380 pKa = 11.33AGVLANLTQSSLNTGIAAGDD400 pKa = 3.73VYY402 pKa = 11.56VSIEE406 pKa = 4.06NLTGSAYY413 pKa = 10.36ADD415 pKa = 3.67SLIGDD420 pKa = 4.31AAANVLKK427 pKa = 10.83GGAGNDD433 pKa = 3.73TLSGGGGSDD442 pKa = 3.49VLIGGIGADD451 pKa = 3.31RR452 pKa = 11.84LNGNGGSDD460 pKa = 3.16TASYY464 pKa = 11.04EE465 pKa = 4.03DD466 pKa = 3.54ATAGVTANLLNASRR480 pKa = 11.84NTGFAAGDD488 pKa = 3.25IYY490 pKa = 11.31GSIEE494 pKa = 4.36HH495 pKa = 6.26LTGSAFDD502 pKa = 5.25DD503 pKa = 4.0LLAGNASANTLRR515 pKa = 11.84GGAGRR520 pKa = 11.84DD521 pKa = 3.63VLSGDD526 pKa = 3.91SGNDD530 pKa = 3.41TLIGGAGADD539 pKa = 3.81KK540 pKa = 10.86LYY542 pKa = 11.25GGAGTDD548 pKa = 3.33LASYY552 pKa = 10.38EE553 pKa = 4.2DD554 pKa = 3.71ATAGVIANLGKK565 pKa = 10.61ASANTGFAAGDD576 pKa = 3.77TYY578 pKa = 11.85ASISGLIGSAFDD590 pKa = 4.05DD591 pKa = 4.09QLTGDD596 pKa = 3.8TGANILIGGNGRR608 pKa = 11.84DD609 pKa = 3.83SLNGGNGNDD618 pKa = 3.79RR619 pKa = 11.84LIGGAGLDD627 pKa = 3.98TLTGGAGADD636 pKa = 3.51TFVYY640 pKa = 10.37AATQEE645 pKa = 4.43SGPSSSARR653 pKa = 11.84DD654 pKa = 3.95LITDD658 pKa = 4.02FSGTSGDD665 pKa = 4.12RR666 pKa = 11.84IDD668 pKa = 4.57VSGIDD673 pKa = 3.55SNLVIGGVQHH683 pKa = 6.63FSLIGGAAFSNTAGQLRR700 pKa = 11.84YY701 pKa = 8.78VTLSTSSFVYY711 pKa = 10.81GDD713 pKa = 3.55TNGDD717 pKa = 3.16GTADD721 pKa = 3.36FTIAFDD727 pKa = 3.91DD728 pKa = 4.05AVTFRR733 pKa = 11.84SDD735 pKa = 3.3YY736 pKa = 11.08FIVV739 pKa = 3.42
MM1 pKa = 7.5IANTGNPLIDD11 pKa = 4.14GLLSGNAWNAATLTYY26 pKa = 10.66AFPTTTSAYY35 pKa = 8.41TYY37 pKa = 10.29TSGTYY42 pKa = 9.77RR43 pKa = 11.84DD44 pKa = 3.78VAPVSLAQQKK54 pKa = 10.11AALFFLEE61 pKa = 4.45QSSGPAANDD70 pKa = 3.56GFSVEE75 pKa = 4.96GFTLLKK81 pKa = 10.06FAAGSATNATLRR93 pKa = 11.84FAEE96 pKa = 4.93SPTADD101 pKa = 3.23PTAYY105 pKa = 10.37AFYY108 pKa = 10.14PGNYY112 pKa = 9.49DD113 pKa = 3.06SAGDD117 pKa = 3.57VWFGTAYY124 pKa = 10.52AGTEE128 pKa = 3.65YY129 pKa = 10.51DD130 pKa = 3.62YY131 pKa = 11.18RR132 pKa = 11.84SPKK135 pKa = 9.36IGTYY139 pKa = 9.13AWHH142 pKa = 6.27TMGHH146 pKa = 6.52EE147 pKa = 4.12IGHH150 pKa = 6.44ALGLKK155 pKa = 9.81HH156 pKa = 5.8GHH158 pKa = 6.35EE159 pKa = 4.67DD160 pKa = 3.13AALPSNVDD168 pKa = 3.4SVEE171 pKa = 3.88YY172 pKa = 10.86SIMTYY177 pKa = 9.9RR178 pKa = 11.84AYY180 pKa = 9.74IGAPLTGYY188 pKa = 10.65SYY190 pKa = 11.62GLNDD194 pKa = 3.76APQSFMSLDD203 pKa = 3.34IAALQAMYY211 pKa = 10.4GADD214 pKa = 3.49YY215 pKa = 10.65TVMNGNTVYY224 pKa = 10.67RR225 pKa = 11.84WTPDD229 pKa = 2.78SGQTIIDD236 pKa = 3.84GVVTIAPGANRR247 pKa = 11.84IFATIWDD254 pKa = 4.11GGGTDD259 pKa = 3.77TFDD262 pKa = 3.19LSAYY266 pKa = 7.55KK267 pKa = 9.52TALSIDD273 pKa = 4.13LQPGKK278 pKa = 10.88YY279 pKa = 9.88SLFSTGQTADD289 pKa = 3.64LGGGPNNGHH298 pKa = 6.24ARR300 pKa = 11.84GNIFNALLYY309 pKa = 10.7KK310 pKa = 10.9NNTASLIEE318 pKa = 4.04NVLGGSGNDD327 pKa = 3.14RR328 pKa = 11.84IVGNQVQNKK337 pKa = 9.57LFGNAGNDD345 pKa = 3.33KK346 pKa = 11.0LFGLTGNDD354 pKa = 2.97ILVGGIGADD363 pKa = 3.53TLDD366 pKa = 4.42GGSGTDD372 pKa = 3.1TASYY376 pKa = 10.32EE377 pKa = 4.03DD378 pKa = 3.78AKK380 pKa = 11.33AGVLANLTQSSLNTGIAAGDD400 pKa = 3.73VYY402 pKa = 11.56VSIEE406 pKa = 4.06NLTGSAYY413 pKa = 10.36ADD415 pKa = 3.67SLIGDD420 pKa = 4.31AAANVLKK427 pKa = 10.83GGAGNDD433 pKa = 3.73TLSGGGGSDD442 pKa = 3.49VLIGGIGADD451 pKa = 3.31RR452 pKa = 11.84LNGNGGSDD460 pKa = 3.16TASYY464 pKa = 11.04EE465 pKa = 4.03DD466 pKa = 3.54ATAGVTANLLNASRR480 pKa = 11.84NTGFAAGDD488 pKa = 3.25IYY490 pKa = 11.31GSIEE494 pKa = 4.36HH495 pKa = 6.26LTGSAFDD502 pKa = 5.25DD503 pKa = 4.0LLAGNASANTLRR515 pKa = 11.84GGAGRR520 pKa = 11.84DD521 pKa = 3.63VLSGDD526 pKa = 3.91SGNDD530 pKa = 3.41TLIGGAGADD539 pKa = 3.81KK540 pKa = 10.86LYY542 pKa = 11.25GGAGTDD548 pKa = 3.33LASYY552 pKa = 10.38EE553 pKa = 4.2DD554 pKa = 3.71ATAGVIANLGKK565 pKa = 10.61ASANTGFAAGDD576 pKa = 3.77TYY578 pKa = 11.85ASISGLIGSAFDD590 pKa = 4.05DD591 pKa = 4.09QLTGDD596 pKa = 3.8TGANILIGGNGRR608 pKa = 11.84DD609 pKa = 3.83SLNGGNGNDD618 pKa = 3.79RR619 pKa = 11.84LIGGAGLDD627 pKa = 3.98TLTGGAGADD636 pKa = 3.51TFVYY640 pKa = 10.37AATQEE645 pKa = 4.43SGPSSSARR653 pKa = 11.84DD654 pKa = 3.95LITDD658 pKa = 4.02FSGTSGDD665 pKa = 4.12RR666 pKa = 11.84IDD668 pKa = 4.57VSGIDD673 pKa = 3.55SNLVIGGVQHH683 pKa = 6.63FSLIGGAAFSNTAGQLRR700 pKa = 11.84YY701 pKa = 8.78VTLSTSSFVYY711 pKa = 10.81GDD713 pKa = 3.55TNGDD717 pKa = 3.16GTADD721 pKa = 3.36FTIAFDD727 pKa = 3.91DD728 pKa = 4.05AVTFRR733 pKa = 11.84SDD735 pKa = 3.3YY736 pKa = 11.08FIVV739 pKa = 3.42
Molecular weight: 74.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5WAD6|A0A0Q5WAD6_9RHIZ Pyridoxamine 5'-phosphate oxidase OS=Rhizobium sp. Leaf371 OX=1736355 GN=ASG39_08540 PE=4 SV=1
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.64GFRR20 pKa = 11.84ARR22 pKa = 11.84IATKK26 pKa = 9.98GGRR29 pKa = 11.84KK30 pKa = 9.73VIVARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.64GFRR20 pKa = 11.84ARR22 pKa = 11.84IATKK26 pKa = 9.98GGRR29 pKa = 11.84KK30 pKa = 9.73VIVARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1421238 |
29 |
3007 |
322.2 |
34.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.398 ± 0.054 | 0.746 ± 0.011 |
5.937 ± 0.032 | 5.518 ± 0.033 |
3.826 ± 0.025 | 8.42 ± 0.036 |
1.976 ± 0.018 | 5.587 ± 0.029 |
3.349 ± 0.036 | 10.047 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.528 ± 0.018 | 2.6 ± 0.021 |
4.949 ± 0.026 | 3.021 ± 0.026 |
6.894 ± 0.04 | 5.672 ± 0.027 |
5.73 ± 0.027 | 7.422 ± 0.029 |
1.198 ± 0.014 | 2.178 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
