
Chlorobium ferrooxidans DSM 13031
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Chlorobi; Chlorobia; Chlorobiales; Chlorobiaceae; Chlorobium/Pelodictyon group; Chlorobium; Chlorobium ferrooxidans
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
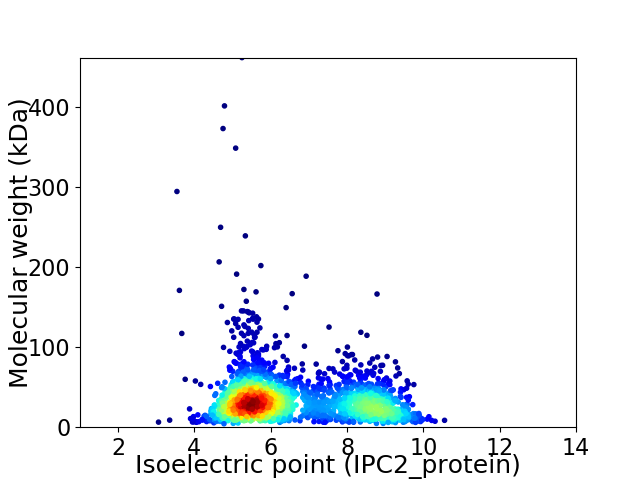
Virtual 2D-PAGE plot for 2158 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
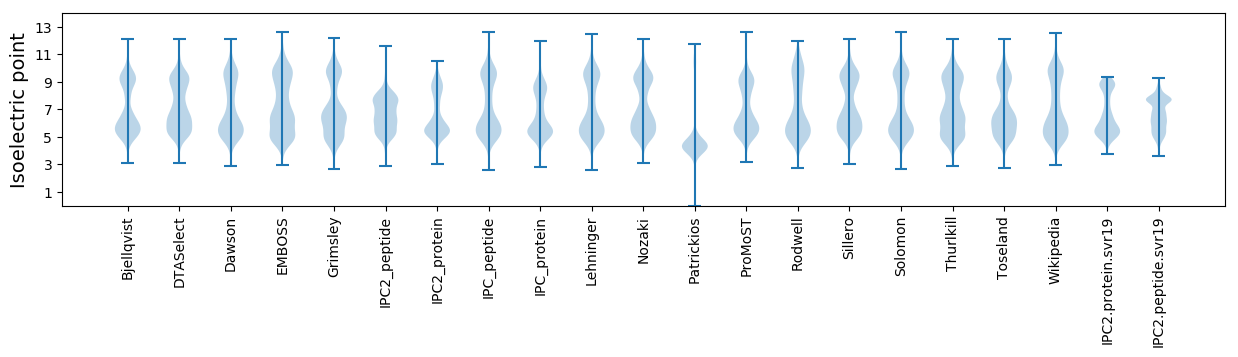
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0YQE8|Q0YQE8_9CHLB PilT protein-like OS=Chlorobium ferrooxidans DSM 13031 OX=377431 GN=CferDRAFT_0590 PE=4 SV=1
MM1 pKa = 7.57IKK3 pKa = 10.6GNEE6 pKa = 4.1SPMSFIKK13 pKa = 10.64QADD16 pKa = 3.66NVGRR20 pKa = 11.84TGTFTIGTNGVWSYY34 pKa = 9.16TANSAFDD41 pKa = 3.58SLNVRR46 pKa = 11.84EE47 pKa = 4.73SIPEE51 pKa = 3.98RR52 pKa = 11.84FSVTLTDD59 pKa = 3.81GKK61 pKa = 8.22TQVEE65 pKa = 4.33RR66 pKa = 11.84VQITGTNDD74 pKa = 2.95AAVISGDD81 pKa = 3.22VSTTIAEE88 pKa = 3.99TDD90 pKa = 3.33EE91 pKa = 4.99ALTATGQLAVTDD103 pKa = 4.01IDD105 pKa = 3.89NPSLFVAQIEE115 pKa = 4.69TEE117 pKa = 4.41GIHH120 pKa = 6.33GKK122 pKa = 10.41FSIDD126 pKa = 3.86EE127 pKa = 4.37SGLWSYY133 pKa = 10.5IANSAFDD140 pKa = 3.98EE141 pKa = 4.51LNEE144 pKa = 4.5GEE146 pKa = 4.67SLTDD150 pKa = 3.5SFIVTAADD158 pKa = 3.36GTTEE162 pKa = 4.14VVRR165 pKa = 11.84VTITGTNEE173 pKa = 3.25ILPAGGSVAEE183 pKa = 4.23SLPVLSSSGNKK194 pKa = 9.22SVAIITGDD202 pKa = 3.19INASALEE209 pKa = 4.16TNAVITATGNLSVASNPLPFVAEE232 pKa = 4.08NGRR235 pKa = 11.84VGSTGIFSIGADD247 pKa = 3.55GLWNFTANSAFDD259 pKa = 3.49GMNVGEE265 pKa = 4.31TVKK268 pKa = 10.84EE269 pKa = 4.18SFTVSTVDD277 pKa = 3.45GMTEE281 pKa = 4.11VVTVTITGTNDD292 pKa = 2.72VAVISGDD299 pKa = 3.51FNATVLEE306 pKa = 4.41TNALIAVTGRR316 pKa = 11.84LSAVDD321 pKa = 3.57IDD323 pKa = 3.85NPAIFSAQSGIKK335 pKa = 10.19GMYY338 pKa = 9.79GVFDD342 pKa = 4.11MGSDD346 pKa = 4.03GAWSYY351 pKa = 11.25RR352 pKa = 11.84AGSAFDD358 pKa = 3.76SLNIGDD364 pKa = 4.5TLTEE368 pKa = 4.26SFTASTIDD376 pKa = 3.57GTSGLVTVTIAGTNDD391 pKa = 3.05AAVITGDD398 pKa = 3.3SSASYY403 pKa = 11.11VEE405 pKa = 4.5TNAAMTVMGRR415 pKa = 11.84LLVNDD420 pKa = 3.9VDD422 pKa = 3.94NPAAFVAQSDD432 pKa = 4.07SKK434 pKa = 11.43GRR436 pKa = 11.84TGTFSIGTDD445 pKa = 3.92GVWSYY450 pKa = 9.11TANSAFDD457 pKa = 3.62GLNVGGKK464 pKa = 9.78VKK466 pKa = 10.8DD467 pKa = 3.72LFSVFTSDD475 pKa = 2.66GTSQVVSVTITGTNDD490 pKa = 2.81AAIITGDD497 pKa = 3.86LSPTVFEE504 pKa = 4.62TNTAVSLTGRR514 pKa = 11.84LRR516 pKa = 11.84AMDD519 pKa = 4.15MDD521 pKa = 4.06NPSIFVAQNGITGLTGTFSIGMDD544 pKa = 3.68GAWSYY549 pKa = 10.86RR550 pKa = 11.84ANSAFDD556 pKa = 3.43SLKK559 pKa = 10.43VGEE562 pKa = 4.6TVTEE566 pKa = 4.23SFSVSTVDD574 pKa = 3.46GTSEE578 pKa = 4.19LVTVIITGTDD588 pKa = 3.31DD589 pKa = 3.71ASVVTGNFSASVAEE603 pKa = 4.01TDD605 pKa = 4.89APITANGTLTVSDD618 pKa = 3.93IDD620 pKa = 3.91SPFAFVAQSGTKK632 pKa = 8.58GTLGLFSIGIDD643 pKa = 3.72GAWDD647 pKa = 4.11FRR649 pKa = 11.84ASSAFDD655 pKa = 3.53NLNVGDD661 pKa = 4.16TLTEE665 pKa = 4.15SFAVTTIDD673 pKa = 3.51GTSDD677 pKa = 3.3LVTVTITGTNDD688 pKa = 2.9AAVITGSLHH697 pKa = 6.39ASEE700 pKa = 5.51DD701 pKa = 3.62EE702 pKa = 4.04SDD704 pKa = 3.99AMITATGTLHH714 pKa = 6.3ATDD717 pKa = 4.39VDD719 pKa = 4.08SSSAFIAQTDD729 pKa = 3.85VAGTNGTFSIGTDD742 pKa = 3.83GVWIFSANSAFDD754 pKa = 3.61GLNVDD759 pKa = 3.83SSVTEE764 pKa = 4.17SFTVTTADD772 pKa = 3.37GTSEE776 pKa = 4.26VVTVTINGTNDD787 pKa = 3.06AAVITGDD794 pKa = 3.96FSATVAEE801 pKa = 4.39TNEE804 pKa = 3.71ASTATGHH811 pKa = 6.28LTVTDD816 pKa = 4.79LDD818 pKa = 3.79NPAIFLIQTDD828 pKa = 3.43NAGFNGSFSIDD839 pKa = 3.44SDD841 pKa = 4.21GLWSYY846 pKa = 10.72SANSAFDD853 pKa = 3.65YY854 pKa = 11.4LNDD857 pKa = 3.72GDD859 pKa = 4.7SLVEE863 pKa = 4.0SFIVEE868 pKa = 4.44SADD871 pKa = 3.31GTSQIVMLTIMGTNDD886 pKa = 3.06AAVITGDD893 pKa = 3.88FSASVDD899 pKa = 3.63EE900 pKa = 4.57TNVPITATGHH910 pKa = 6.63LSVTDD915 pKa = 3.46VDD917 pKa = 4.12NPALFVVQTDD927 pKa = 3.35TGGINGSFSIDD938 pKa = 3.33AAGLWSYY945 pKa = 10.68AANSAFDD952 pKa = 4.0EE953 pKa = 4.59LNAGDD958 pKa = 4.29TLTEE962 pKa = 4.18SFTVSTMDD970 pKa = 3.02GTTEE974 pKa = 3.8IVTVTITGTNDD985 pKa = 3.11ALTTSQIVTQLTTSWGSGLTGYY1007 pKa = 8.51TFTWPADD1014 pKa = 3.66LTSISYY1020 pKa = 10.32AINEE1024 pKa = 4.23GTPTNVDD1031 pKa = 3.08GYY1033 pKa = 8.71TPSEE1037 pKa = 4.37GGTSLVQMSALQVATARR1054 pKa = 11.84LSFQLLDD1061 pKa = 4.87DD1062 pKa = 4.37IMAKK1066 pKa = 10.29SSGADD1071 pKa = 3.12HH1072 pKa = 7.17RR1073 pKa = 11.84LVQIADD1079 pKa = 3.62PSANITLNYY1088 pKa = 9.95SSNTGGGTYY1097 pKa = 9.7TISFGTVDD1105 pKa = 3.41TVTQSFSITAEE1116 pKa = 4.17QIWLSSAWSSNSDD1129 pKa = 2.43IGMSLGSYY1137 pKa = 10.78GLFTMIHH1144 pKa = 6.58EE1145 pKa = 4.8IGHH1148 pKa = 6.56ALGLSHH1154 pKa = 7.48PGVYY1158 pKa = 10.16DD1159 pKa = 3.71AGNGGTITYY1168 pKa = 10.13QNNAAFAEE1176 pKa = 4.47DD1177 pKa = 3.41NRR1179 pKa = 11.84KK1180 pKa = 7.38YY1181 pKa = 9.59TVMSYY1186 pKa = 10.51FGGYY1190 pKa = 9.51LPGYY1194 pKa = 8.75GWYY1197 pKa = 9.43PDD1199 pKa = 3.39GTYY1202 pKa = 10.92SSFIFPQTPMLYY1214 pKa = 10.32DD1215 pKa = 3.65IAAIQALHH1223 pKa = 6.41GADD1226 pKa = 3.0TVTRR1230 pKa = 11.84TGDD1233 pKa = 3.3TIYY1236 pKa = 11.09GFNCNLASTDD1246 pKa = 3.56AEE1248 pKa = 3.95RR1249 pKa = 11.84PIYY1252 pKa = 10.68DD1253 pKa = 3.62FTSNATPIFTIWDD1266 pKa = 3.69AGGMDD1271 pKa = 4.58TIDD1274 pKa = 3.43CSGYY1278 pKa = 6.87TTQQIINLNPGTYY1291 pKa = 9.97SSLGGMTEE1299 pKa = 3.93NVAIAFDD1306 pKa = 4.03CTIEE1310 pKa = 4.09RR1311 pKa = 11.84AVGGSGNDD1319 pKa = 3.44VLIGSAALNYY1329 pKa = 10.64LNGGNAGDD1337 pKa = 3.99FYY1339 pKa = 10.91IVNRR1343 pKa = 11.84AGDD1346 pKa = 3.55HH1347 pKa = 6.06LAAEE1351 pKa = 4.71FADD1354 pKa = 3.82TGTTGSDD1361 pKa = 3.03EE1362 pKa = 4.07VRR1364 pKa = 11.84FASTIEE1370 pKa = 4.22HH1371 pKa = 6.67DD1372 pKa = 4.05TLTLFAGDD1380 pKa = 3.53TGIEE1384 pKa = 3.85RR1385 pKa = 11.84VVIGTGVGISGVTTGTTSLDD1405 pKa = 3.13VDD1407 pKa = 4.12ASAVLNPIVITGNAGANKK1425 pKa = 8.89LTGTTYY1431 pKa = 11.5NDD1433 pKa = 2.92VLYY1436 pKa = 10.56GGKK1439 pKa = 8.34GTDD1442 pKa = 4.19LLNGGNGSDD1451 pKa = 3.23IYY1453 pKa = 10.85HH1454 pKa = 6.87IFSADD1459 pKa = 3.07EE1460 pKa = 3.96HH1461 pKa = 7.15LSAEE1465 pKa = 4.36FSDD1468 pKa = 3.78TGTTGSDD1475 pKa = 3.11DD1476 pKa = 3.49VRR1478 pKa = 11.84FGSTIANDD1486 pKa = 3.81TLTLYY1491 pKa = 10.9AGDD1494 pKa = 3.73TGIEE1498 pKa = 4.01RR1499 pKa = 11.84VTATGTATLNIDD1511 pKa = 3.37ASGVLNGLLLTGNAGSNTLTGTAYY1535 pKa = 10.93CDD1537 pKa = 3.46TFTGGSGADD1546 pKa = 3.28SFVFTYY1552 pKa = 10.49QLQPVVNKK1560 pKa = 9.12DD1561 pKa = 3.54TIIDD1565 pKa = 3.9FQSGSDD1571 pKa = 3.32VLAFSKK1577 pKa = 11.03GSFTGITSSAGGLLSSAEE1595 pKa = 4.11FWSEE1599 pKa = 3.29AGATVGHH1606 pKa = 7.13DD1607 pKa = 3.17SDD1609 pKa = 5.57DD1610 pKa = 4.69RR1611 pKa = 11.84IIYY1614 pKa = 7.58DD1615 pKa = 3.59TATGNLYY1622 pKa = 10.79YY1623 pKa = 10.61DD1624 pKa = 4.38DD1625 pKa = 5.87DD1626 pKa = 5.74GNGSANARR1634 pKa = 11.84LVALIGTTAHH1644 pKa = 7.04PALLYY1649 pKa = 10.12TDD1651 pKa = 3.98ISIISS1656 pKa = 3.46
MM1 pKa = 7.57IKK3 pKa = 10.6GNEE6 pKa = 4.1SPMSFIKK13 pKa = 10.64QADD16 pKa = 3.66NVGRR20 pKa = 11.84TGTFTIGTNGVWSYY34 pKa = 9.16TANSAFDD41 pKa = 3.58SLNVRR46 pKa = 11.84EE47 pKa = 4.73SIPEE51 pKa = 3.98RR52 pKa = 11.84FSVTLTDD59 pKa = 3.81GKK61 pKa = 8.22TQVEE65 pKa = 4.33RR66 pKa = 11.84VQITGTNDD74 pKa = 2.95AAVISGDD81 pKa = 3.22VSTTIAEE88 pKa = 3.99TDD90 pKa = 3.33EE91 pKa = 4.99ALTATGQLAVTDD103 pKa = 4.01IDD105 pKa = 3.89NPSLFVAQIEE115 pKa = 4.69TEE117 pKa = 4.41GIHH120 pKa = 6.33GKK122 pKa = 10.41FSIDD126 pKa = 3.86EE127 pKa = 4.37SGLWSYY133 pKa = 10.5IANSAFDD140 pKa = 3.98EE141 pKa = 4.51LNEE144 pKa = 4.5GEE146 pKa = 4.67SLTDD150 pKa = 3.5SFIVTAADD158 pKa = 3.36GTTEE162 pKa = 4.14VVRR165 pKa = 11.84VTITGTNEE173 pKa = 3.25ILPAGGSVAEE183 pKa = 4.23SLPVLSSSGNKK194 pKa = 9.22SVAIITGDD202 pKa = 3.19INASALEE209 pKa = 4.16TNAVITATGNLSVASNPLPFVAEE232 pKa = 4.08NGRR235 pKa = 11.84VGSTGIFSIGADD247 pKa = 3.55GLWNFTANSAFDD259 pKa = 3.49GMNVGEE265 pKa = 4.31TVKK268 pKa = 10.84EE269 pKa = 4.18SFTVSTVDD277 pKa = 3.45GMTEE281 pKa = 4.11VVTVTITGTNDD292 pKa = 2.72VAVISGDD299 pKa = 3.51FNATVLEE306 pKa = 4.41TNALIAVTGRR316 pKa = 11.84LSAVDD321 pKa = 3.57IDD323 pKa = 3.85NPAIFSAQSGIKK335 pKa = 10.19GMYY338 pKa = 9.79GVFDD342 pKa = 4.11MGSDD346 pKa = 4.03GAWSYY351 pKa = 11.25RR352 pKa = 11.84AGSAFDD358 pKa = 3.76SLNIGDD364 pKa = 4.5TLTEE368 pKa = 4.26SFTASTIDD376 pKa = 3.57GTSGLVTVTIAGTNDD391 pKa = 3.05AAVITGDD398 pKa = 3.3SSASYY403 pKa = 11.11VEE405 pKa = 4.5TNAAMTVMGRR415 pKa = 11.84LLVNDD420 pKa = 3.9VDD422 pKa = 3.94NPAAFVAQSDD432 pKa = 4.07SKK434 pKa = 11.43GRR436 pKa = 11.84TGTFSIGTDD445 pKa = 3.92GVWSYY450 pKa = 9.11TANSAFDD457 pKa = 3.62GLNVGGKK464 pKa = 9.78VKK466 pKa = 10.8DD467 pKa = 3.72LFSVFTSDD475 pKa = 2.66GTSQVVSVTITGTNDD490 pKa = 2.81AAIITGDD497 pKa = 3.86LSPTVFEE504 pKa = 4.62TNTAVSLTGRR514 pKa = 11.84LRR516 pKa = 11.84AMDD519 pKa = 4.15MDD521 pKa = 4.06NPSIFVAQNGITGLTGTFSIGMDD544 pKa = 3.68GAWSYY549 pKa = 10.86RR550 pKa = 11.84ANSAFDD556 pKa = 3.43SLKK559 pKa = 10.43VGEE562 pKa = 4.6TVTEE566 pKa = 4.23SFSVSTVDD574 pKa = 3.46GTSEE578 pKa = 4.19LVTVIITGTDD588 pKa = 3.31DD589 pKa = 3.71ASVVTGNFSASVAEE603 pKa = 4.01TDD605 pKa = 4.89APITANGTLTVSDD618 pKa = 3.93IDD620 pKa = 3.91SPFAFVAQSGTKK632 pKa = 8.58GTLGLFSIGIDD643 pKa = 3.72GAWDD647 pKa = 4.11FRR649 pKa = 11.84ASSAFDD655 pKa = 3.53NLNVGDD661 pKa = 4.16TLTEE665 pKa = 4.15SFAVTTIDD673 pKa = 3.51GTSDD677 pKa = 3.3LVTVTITGTNDD688 pKa = 2.9AAVITGSLHH697 pKa = 6.39ASEE700 pKa = 5.51DD701 pKa = 3.62EE702 pKa = 4.04SDD704 pKa = 3.99AMITATGTLHH714 pKa = 6.3ATDD717 pKa = 4.39VDD719 pKa = 4.08SSSAFIAQTDD729 pKa = 3.85VAGTNGTFSIGTDD742 pKa = 3.83GVWIFSANSAFDD754 pKa = 3.61GLNVDD759 pKa = 3.83SSVTEE764 pKa = 4.17SFTVTTADD772 pKa = 3.37GTSEE776 pKa = 4.26VVTVTINGTNDD787 pKa = 3.06AAVITGDD794 pKa = 3.96FSATVAEE801 pKa = 4.39TNEE804 pKa = 3.71ASTATGHH811 pKa = 6.28LTVTDD816 pKa = 4.79LDD818 pKa = 3.79NPAIFLIQTDD828 pKa = 3.43NAGFNGSFSIDD839 pKa = 3.44SDD841 pKa = 4.21GLWSYY846 pKa = 10.72SANSAFDD853 pKa = 3.65YY854 pKa = 11.4LNDD857 pKa = 3.72GDD859 pKa = 4.7SLVEE863 pKa = 4.0SFIVEE868 pKa = 4.44SADD871 pKa = 3.31GTSQIVMLTIMGTNDD886 pKa = 3.06AAVITGDD893 pKa = 3.88FSASVDD899 pKa = 3.63EE900 pKa = 4.57TNVPITATGHH910 pKa = 6.63LSVTDD915 pKa = 3.46VDD917 pKa = 4.12NPALFVVQTDD927 pKa = 3.35TGGINGSFSIDD938 pKa = 3.33AAGLWSYY945 pKa = 10.68AANSAFDD952 pKa = 4.0EE953 pKa = 4.59LNAGDD958 pKa = 4.29TLTEE962 pKa = 4.18SFTVSTMDD970 pKa = 3.02GTTEE974 pKa = 3.8IVTVTITGTNDD985 pKa = 3.11ALTTSQIVTQLTTSWGSGLTGYY1007 pKa = 8.51TFTWPADD1014 pKa = 3.66LTSISYY1020 pKa = 10.32AINEE1024 pKa = 4.23GTPTNVDD1031 pKa = 3.08GYY1033 pKa = 8.71TPSEE1037 pKa = 4.37GGTSLVQMSALQVATARR1054 pKa = 11.84LSFQLLDD1061 pKa = 4.87DD1062 pKa = 4.37IMAKK1066 pKa = 10.29SSGADD1071 pKa = 3.12HH1072 pKa = 7.17RR1073 pKa = 11.84LVQIADD1079 pKa = 3.62PSANITLNYY1088 pKa = 9.95SSNTGGGTYY1097 pKa = 9.7TISFGTVDD1105 pKa = 3.41TVTQSFSITAEE1116 pKa = 4.17QIWLSSAWSSNSDD1129 pKa = 2.43IGMSLGSYY1137 pKa = 10.78GLFTMIHH1144 pKa = 6.58EE1145 pKa = 4.8IGHH1148 pKa = 6.56ALGLSHH1154 pKa = 7.48PGVYY1158 pKa = 10.16DD1159 pKa = 3.71AGNGGTITYY1168 pKa = 10.13QNNAAFAEE1176 pKa = 4.47DD1177 pKa = 3.41NRR1179 pKa = 11.84KK1180 pKa = 7.38YY1181 pKa = 9.59TVMSYY1186 pKa = 10.51FGGYY1190 pKa = 9.51LPGYY1194 pKa = 8.75GWYY1197 pKa = 9.43PDD1199 pKa = 3.39GTYY1202 pKa = 10.92SSFIFPQTPMLYY1214 pKa = 10.32DD1215 pKa = 3.65IAAIQALHH1223 pKa = 6.41GADD1226 pKa = 3.0TVTRR1230 pKa = 11.84TGDD1233 pKa = 3.3TIYY1236 pKa = 11.09GFNCNLASTDD1246 pKa = 3.56AEE1248 pKa = 3.95RR1249 pKa = 11.84PIYY1252 pKa = 10.68DD1253 pKa = 3.62FTSNATPIFTIWDD1266 pKa = 3.69AGGMDD1271 pKa = 4.58TIDD1274 pKa = 3.43CSGYY1278 pKa = 6.87TTQQIINLNPGTYY1291 pKa = 9.97SSLGGMTEE1299 pKa = 3.93NVAIAFDD1306 pKa = 4.03CTIEE1310 pKa = 4.09RR1311 pKa = 11.84AVGGSGNDD1319 pKa = 3.44VLIGSAALNYY1329 pKa = 10.64LNGGNAGDD1337 pKa = 3.99FYY1339 pKa = 10.91IVNRR1343 pKa = 11.84AGDD1346 pKa = 3.55HH1347 pKa = 6.06LAAEE1351 pKa = 4.71FADD1354 pKa = 3.82TGTTGSDD1361 pKa = 3.03EE1362 pKa = 4.07VRR1364 pKa = 11.84FASTIEE1370 pKa = 4.22HH1371 pKa = 6.67DD1372 pKa = 4.05TLTLFAGDD1380 pKa = 3.53TGIEE1384 pKa = 3.85RR1385 pKa = 11.84VVIGTGVGISGVTTGTTSLDD1405 pKa = 3.13VDD1407 pKa = 4.12ASAVLNPIVITGNAGANKK1425 pKa = 8.89LTGTTYY1431 pKa = 11.5NDD1433 pKa = 2.92VLYY1436 pKa = 10.56GGKK1439 pKa = 8.34GTDD1442 pKa = 4.19LLNGGNGSDD1451 pKa = 3.23IYY1453 pKa = 10.85HH1454 pKa = 6.87IFSADD1459 pKa = 3.07EE1460 pKa = 3.96HH1461 pKa = 7.15LSAEE1465 pKa = 4.36FSDD1468 pKa = 3.78TGTTGSDD1475 pKa = 3.11DD1476 pKa = 3.49VRR1478 pKa = 11.84FGSTIANDD1486 pKa = 3.81TLTLYY1491 pKa = 10.9AGDD1494 pKa = 3.73TGIEE1498 pKa = 4.01RR1499 pKa = 11.84VTATGTATLNIDD1511 pKa = 3.37ASGVLNGLLLTGNAGSNTLTGTAYY1535 pKa = 10.93CDD1537 pKa = 3.46TFTGGSGADD1546 pKa = 3.28SFVFTYY1552 pKa = 10.49QLQPVVNKK1560 pKa = 9.12DD1561 pKa = 3.54TIIDD1565 pKa = 3.9FQSGSDD1571 pKa = 3.32VLAFSKK1577 pKa = 11.03GSFTGITSSAGGLLSSAEE1595 pKa = 4.11FWSEE1599 pKa = 3.29AGATVGHH1606 pKa = 7.13DD1607 pKa = 3.17SDD1609 pKa = 5.57DD1610 pKa = 4.69RR1611 pKa = 11.84IIYY1614 pKa = 7.58DD1615 pKa = 3.59TATGNLYY1622 pKa = 10.79YY1623 pKa = 10.61DD1624 pKa = 4.38DD1625 pKa = 5.87DD1626 pKa = 5.74GNGSANARR1634 pKa = 11.84LVALIGTTAHH1644 pKa = 7.04PALLYY1649 pKa = 10.12TDD1651 pKa = 3.98ISIISS1656 pKa = 3.46
Molecular weight: 170.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0YRE3|Q0YRE3_9CHLB Cyclic peptide transporter OS=Chlorobium ferrooxidans DSM 13031 OX=377431 GN=CferDRAFT_0891 PE=4 SV=1
MM1 pKa = 7.45LLSRR5 pKa = 11.84VLPPLQVWKK14 pKa = 9.29TEE16 pKa = 3.7AMKK19 pKa = 10.63QSSTPRR25 pKa = 11.84VCRR28 pKa = 11.84DD29 pKa = 2.78NSFTSKK35 pKa = 10.23QPQKK39 pKa = 10.7ARR41 pKa = 11.84LRR43 pKa = 11.84IVLFLLVIPLLLALSLLFGPSGIGLPDD70 pKa = 3.47MNSPAGGAILSLRR83 pKa = 11.84YY84 pKa = 9.65SRR86 pKa = 11.84LLMGMMTGAALSASGVVFQALLRR109 pKa = 11.84NPLAEE114 pKa = 4.37PYY116 pKa = 10.58VLGVSGGAGLGATISILAGSSVILSLSLPVVAFVSAVLTLMVVYY160 pKa = 9.7GIARR164 pKa = 11.84QGSSGQPSVYY174 pKa = 10.69SLILSGVIVSSICSSIIMFMVSTASVEE201 pKa = 4.17GLHH204 pKa = 6.58NVIWWMLGSLQPVSTGQQLLSLGLIVSALFGIWLLSPRR242 pKa = 11.84LNALALGRR250 pKa = 11.84EE251 pKa = 4.05MAHH254 pKa = 5.38YY255 pKa = 10.17QGVHH259 pKa = 6.29ADD261 pKa = 4.42LIIVSGLLCATLLAATAVSLSGMIGFVGLIVPHH294 pKa = 6.08VMRR297 pKa = 11.84ALFGPDD303 pKa = 2.84NRR305 pKa = 11.84RR306 pKa = 11.84LVPLSALGGGTFLVLCDD323 pKa = 3.52AVARR327 pKa = 11.84TLMAPVEE334 pKa = 4.25IPVGVVTALAGGPFFLIILQRR355 pKa = 11.84RR356 pKa = 11.84MKK358 pKa = 9.9QAWIAA363 pKa = 3.37
MM1 pKa = 7.45LLSRR5 pKa = 11.84VLPPLQVWKK14 pKa = 9.29TEE16 pKa = 3.7AMKK19 pKa = 10.63QSSTPRR25 pKa = 11.84VCRR28 pKa = 11.84DD29 pKa = 2.78NSFTSKK35 pKa = 10.23QPQKK39 pKa = 10.7ARR41 pKa = 11.84LRR43 pKa = 11.84IVLFLLVIPLLLALSLLFGPSGIGLPDD70 pKa = 3.47MNSPAGGAILSLRR83 pKa = 11.84YY84 pKa = 9.65SRR86 pKa = 11.84LLMGMMTGAALSASGVVFQALLRR109 pKa = 11.84NPLAEE114 pKa = 4.37PYY116 pKa = 10.58VLGVSGGAGLGATISILAGSSVILSLSLPVVAFVSAVLTLMVVYY160 pKa = 9.7GIARR164 pKa = 11.84QGSSGQPSVYY174 pKa = 10.69SLILSGVIVSSICSSIIMFMVSTASVEE201 pKa = 4.17GLHH204 pKa = 6.58NVIWWMLGSLQPVSTGQQLLSLGLIVSALFGIWLLSPRR242 pKa = 11.84LNALALGRR250 pKa = 11.84EE251 pKa = 4.05MAHH254 pKa = 5.38YY255 pKa = 10.17QGVHH259 pKa = 6.29ADD261 pKa = 4.42LIIVSGLLCATLLAATAVSLSGMIGFVGLIVPHH294 pKa = 6.08VMRR297 pKa = 11.84ALFGPDD303 pKa = 2.84NRR305 pKa = 11.84RR306 pKa = 11.84LVPLSALGGGTFLVLCDD323 pKa = 3.52AVARR327 pKa = 11.84TLMAPVEE334 pKa = 4.25IPVGVVTALAGGPFFLIILQRR355 pKa = 11.84RR356 pKa = 11.84MKK358 pKa = 9.9QAWIAA363 pKa = 3.37
Molecular weight: 38.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
717850 |
39 |
4189 |
332.6 |
36.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.715 ± 0.059 | 1.057 ± 0.021 |
5.027 ± 0.042 | 6.666 ± 0.072 |
4.333 ± 0.045 | 7.446 ± 0.096 |
2.131 ± 0.03 | 6.688 ± 0.051 |
5.17 ± 0.065 | 10.386 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.429 ± 0.03 | 3.717 ± 0.042 |
4.194 ± 0.043 | 3.145 ± 0.033 |
5.664 ± 0.067 | 7.046 ± 0.073 |
5.424 ± 0.119 | 6.723 ± 0.042 |
1.024 ± 0.021 | 3.014 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
