
Burkholderiales bacterium LSUCC0115
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; unclassified Burkholderiales
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
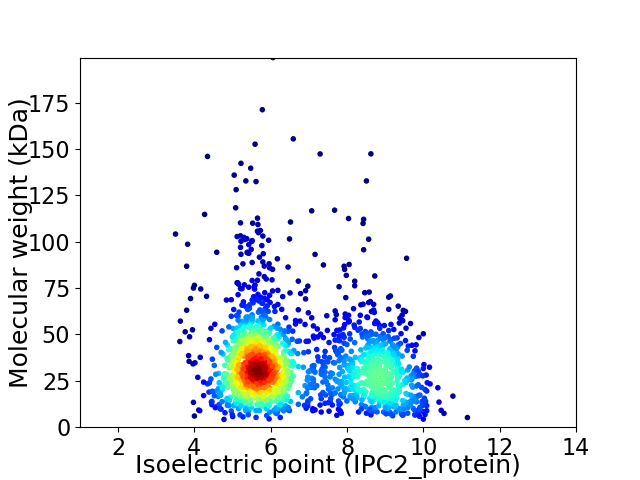
Virtual 2D-PAGE plot for 1762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7C9GS29|A0A7C9GS29_9BURK Exodeoxyribonuclease III OS=Burkholderiales bacterium LSUCC0115 OX=1778438 GN=xth PE=3 SV=1
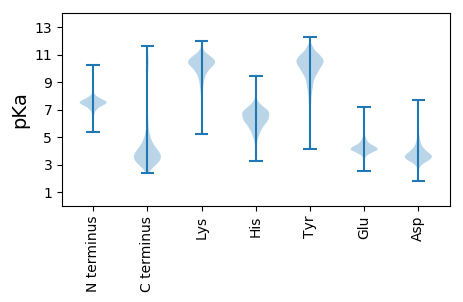
MM1 pKa = 7.4TLRR4 pKa = 11.84VYY6 pKa = 10.18YY7 pKa = 11.08VNGFDD12 pKa = 4.43EE13 pKa = 6.17GEE15 pKa = 4.14DD16 pKa = 3.6PQYY19 pKa = 11.56VGLEE23 pKa = 3.73HH24 pKa = 7.45AIYY27 pKa = 10.89YY28 pKa = 9.97SDD30 pKa = 4.04RR31 pKa = 11.84NSNTGTWSDD40 pKa = 3.44RR41 pKa = 11.84APIKK45 pKa = 10.54IEE47 pKa = 3.82GQIDD51 pKa = 3.81GNAVDD56 pKa = 4.52PNLIQLEE63 pKa = 4.22DD64 pKa = 3.35GRR66 pKa = 11.84YY67 pKa = 9.49LLTYY71 pKa = 9.71MRR73 pKa = 11.84GQFTQTNPEE82 pKa = 4.18PVSTIYY88 pKa = 10.7SAWSYY93 pKa = 11.48NGVDD97 pKa = 4.26FYY99 pKa = 11.84NPQVLYY105 pKa = 10.4KK106 pKa = 9.86PSSQASFGLPAVTDD120 pKa = 3.97PSLVQLTDD128 pKa = 3.87GSWLLAISNPSSQSAGLYY146 pKa = 8.54TSSDD150 pKa = 3.21GVNFTANGVSLPAFSPDD167 pKa = 3.28FQLLANGNVRR177 pKa = 11.84IVYY180 pKa = 10.41ADD182 pKa = 3.46AAAGGFGSQVSSDD195 pKa = 3.65GGEE198 pKa = 4.0TWVVEE203 pKa = 4.31SGARR207 pKa = 11.84FAGPYY212 pKa = 9.42FDD214 pKa = 4.69PSVFRR219 pKa = 11.84MPDD222 pKa = 3.3GSWGMFYY229 pKa = 9.36KK230 pKa = 9.95TQTTVEE236 pKa = 4.41GQQSSPLLGHH246 pKa = 6.73LTSLATSSDD255 pKa = 2.9GTTFTTQDD263 pKa = 3.12AEE265 pKa = 4.23FAAAASVAEE274 pKa = 5.14GIDD277 pKa = 3.93FSGLTVSEE285 pKa = 4.23SLIAATSTNDD295 pKa = 3.5TIAVGEE301 pKa = 4.15QGAHH305 pKa = 3.87VRR307 pKa = 11.84AGNGIDD313 pKa = 3.32TVSFAIASSALTLEE327 pKa = 3.63QDD329 pKa = 4.53FFNEE333 pKa = 4.15GDD335 pKa = 3.6WVGHH339 pKa = 6.23LLSDD343 pKa = 4.16ANVAYY348 pKa = 10.72LLQDD352 pKa = 2.88VEE354 pKa = 4.99RR355 pKa = 11.84FVFTDD360 pKa = 3.1KK361 pKa = 11.2SLAVDD366 pKa = 4.37LDD368 pKa = 3.7GHH370 pKa = 6.68AGQVAKK376 pKa = 10.67ILGAVFGSDD385 pKa = 2.68SVANAEE391 pKa = 4.18YY392 pKa = 10.89VGIGLDD398 pKa = 3.73LLDD401 pKa = 5.07GGMSYY406 pKa = 11.48SDD408 pKa = 3.83LAALAVSVTGNTSSTDD424 pKa = 2.75ICNLLWEE431 pKa = 4.35NVIGSSATATDD442 pKa = 3.51IAPFKK447 pKa = 11.31SMLDD451 pKa = 3.42TGQLSIGQLTTLAADD466 pKa = 3.67TSFNTDD472 pKa = 3.13NIDD475 pKa = 3.62LVGLSEE481 pKa = 4.91IGLEE485 pKa = 4.63FII487 pKa = 5.54
MM1 pKa = 7.4TLRR4 pKa = 11.84VYY6 pKa = 10.18YY7 pKa = 11.08VNGFDD12 pKa = 4.43EE13 pKa = 6.17GEE15 pKa = 4.14DD16 pKa = 3.6PQYY19 pKa = 11.56VGLEE23 pKa = 3.73HH24 pKa = 7.45AIYY27 pKa = 10.89YY28 pKa = 9.97SDD30 pKa = 4.04RR31 pKa = 11.84NSNTGTWSDD40 pKa = 3.44RR41 pKa = 11.84APIKK45 pKa = 10.54IEE47 pKa = 3.82GQIDD51 pKa = 3.81GNAVDD56 pKa = 4.52PNLIQLEE63 pKa = 4.22DD64 pKa = 3.35GRR66 pKa = 11.84YY67 pKa = 9.49LLTYY71 pKa = 9.71MRR73 pKa = 11.84GQFTQTNPEE82 pKa = 4.18PVSTIYY88 pKa = 10.7SAWSYY93 pKa = 11.48NGVDD97 pKa = 4.26FYY99 pKa = 11.84NPQVLYY105 pKa = 10.4KK106 pKa = 9.86PSSQASFGLPAVTDD120 pKa = 3.97PSLVQLTDD128 pKa = 3.87GSWLLAISNPSSQSAGLYY146 pKa = 8.54TSSDD150 pKa = 3.21GVNFTANGVSLPAFSPDD167 pKa = 3.28FQLLANGNVRR177 pKa = 11.84IVYY180 pKa = 10.41ADD182 pKa = 3.46AAAGGFGSQVSSDD195 pKa = 3.65GGEE198 pKa = 4.0TWVVEE203 pKa = 4.31SGARR207 pKa = 11.84FAGPYY212 pKa = 9.42FDD214 pKa = 4.69PSVFRR219 pKa = 11.84MPDD222 pKa = 3.3GSWGMFYY229 pKa = 9.36KK230 pKa = 9.95TQTTVEE236 pKa = 4.41GQQSSPLLGHH246 pKa = 6.73LTSLATSSDD255 pKa = 2.9GTTFTTQDD263 pKa = 3.12AEE265 pKa = 4.23FAAAASVAEE274 pKa = 5.14GIDD277 pKa = 3.93FSGLTVSEE285 pKa = 4.23SLIAATSTNDD295 pKa = 3.5TIAVGEE301 pKa = 4.15QGAHH305 pKa = 3.87VRR307 pKa = 11.84AGNGIDD313 pKa = 3.32TVSFAIASSALTLEE327 pKa = 3.63QDD329 pKa = 4.53FFNEE333 pKa = 4.15GDD335 pKa = 3.6WVGHH339 pKa = 6.23LLSDD343 pKa = 4.16ANVAYY348 pKa = 10.72LLQDD352 pKa = 2.88VEE354 pKa = 4.99RR355 pKa = 11.84FVFTDD360 pKa = 3.1KK361 pKa = 11.2SLAVDD366 pKa = 4.37LDD368 pKa = 3.7GHH370 pKa = 6.68AGQVAKK376 pKa = 10.67ILGAVFGSDD385 pKa = 2.68SVANAEE391 pKa = 4.18YY392 pKa = 10.89VGIGLDD398 pKa = 3.73LLDD401 pKa = 5.07GGMSYY406 pKa = 11.48SDD408 pKa = 3.83LAALAVSVTGNTSSTDD424 pKa = 2.75ICNLLWEE431 pKa = 4.35NVIGSSATATDD442 pKa = 3.51IAPFKK447 pKa = 11.31SMLDD451 pKa = 3.42TGQLSIGQLTTLAADD466 pKa = 3.67TSFNTDD472 pKa = 3.13NIDD475 pKa = 3.62LVGLSEE481 pKa = 4.91IGLEE485 pKa = 4.63FII487 pKa = 5.54
Molecular weight: 51.43 kDa
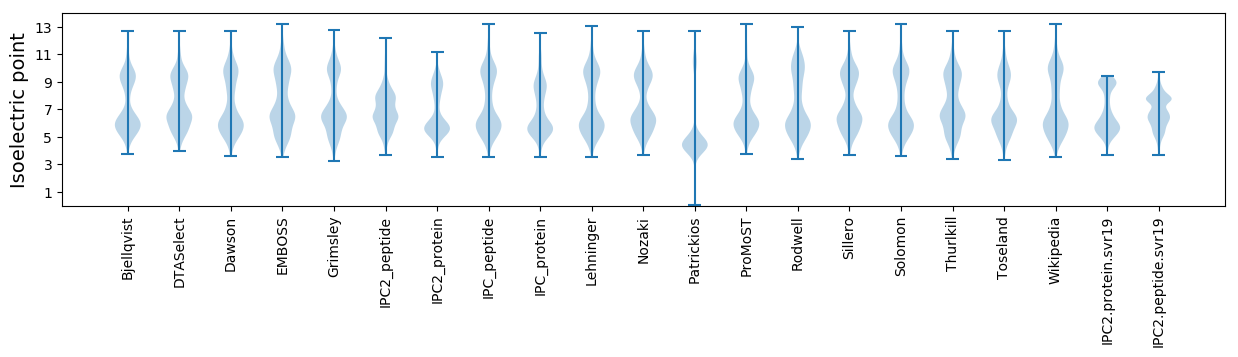
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7C9GTN5|A0A7C9GTN5_9BURK Uncharacterized protein OS=Burkholderiales bacterium LSUCC0115 OX=1778438 GN=GH816_07535 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.67GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.67GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
579727 |
37 |
1818 |
329.0 |
35.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.457 ± 0.06 | 0.952 ± 0.021 |
5.311 ± 0.044 | 5.143 ± 0.052 |
3.6 ± 0.04 | 8.177 ± 0.05 |
2.148 ± 0.03 | 4.834 ± 0.044 |
3.869 ± 0.055 | 10.858 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.648 ± 0.027 | 2.661 ± 0.033 |
4.952 ± 0.039 | 4.827 ± 0.045 |
6.132 ± 0.049 | 6.156 ± 0.05 |
5.061 ± 0.043 | 7.623 ± 0.046 |
1.426 ± 0.027 | 2.163 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
