
Hubei sobemo-like virus 23
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
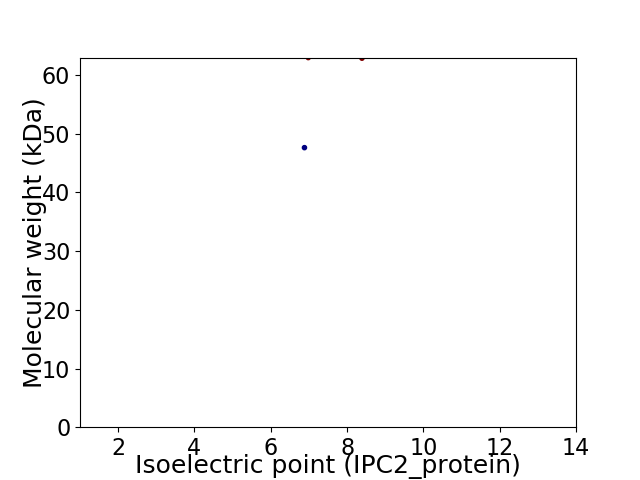
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
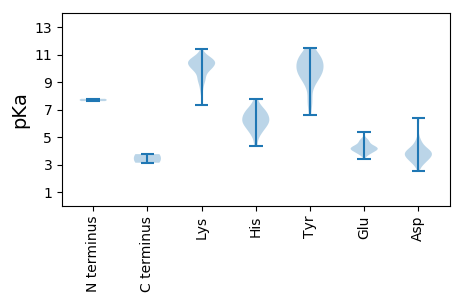
Protein with the lowest isoelectric point:
>tr|A0A1L3KEQ3|A0A1L3KEQ3_9VIRU C2H2-type domain-containing protein OS=Hubei sobemo-like virus 23 OX=1923209 PE=4 SV=1
MM1 pKa = 7.61ASDD4 pKa = 4.02FTWPLLGGDD13 pKa = 3.71AEE15 pKa = 4.42LRR17 pKa = 11.84SLLTHH22 pKa = 5.46VQMSEE27 pKa = 3.76GLRR30 pKa = 11.84EE31 pKa = 4.77DD32 pKa = 4.58GPSHH36 pKa = 6.42QIKK39 pKa = 9.16TKK41 pKa = 10.58LLDD44 pKa = 3.3IAEE47 pKa = 4.84DD48 pKa = 3.58MFKK51 pKa = 10.16PARR54 pKa = 11.84WRR56 pKa = 11.84IPDD59 pKa = 3.93DD60 pKa = 3.57FLSFNHH66 pKa = 6.07YY67 pKa = 9.38MRR69 pKa = 11.84TLNQLVWTSSPGLPYY84 pKa = 9.93MRR86 pKa = 11.84TYY88 pKa = 10.39PNNGGMFGVVDD99 pKa = 4.28GVPNPEE105 pKa = 3.88KK106 pKa = 10.5CRR108 pKa = 11.84YY109 pKa = 6.63WWTIVQQKK117 pKa = 9.68IRR119 pKa = 11.84DD120 pKa = 3.67QFSDD124 pKa = 3.82YY125 pKa = 10.71IRR127 pKa = 11.84VFIKK131 pKa = 10.33QEE133 pKa = 4.12PITRR137 pKa = 11.84KK138 pKa = 9.98KK139 pKa = 10.57FEE141 pKa = 4.07QNRR144 pKa = 11.84FRR146 pKa = 11.84LIFSVSVLDD155 pKa = 4.56QIIDD159 pKa = 3.35HH160 pKa = 6.2MLFGDD165 pKa = 4.09FNDD168 pKa = 3.94LVIEE172 pKa = 3.99NWYY175 pKa = 10.19NLPTKK180 pKa = 10.53VGWSPLGGGWKK191 pKa = 9.93IIPKK195 pKa = 9.31SKK197 pKa = 10.2CHH199 pKa = 6.93AIDD202 pKa = 3.1KK203 pKa = 10.08RR204 pKa = 11.84AWDD207 pKa = 3.59WTVKK211 pKa = 8.5MWLIEE216 pKa = 4.19LEE218 pKa = 3.87FMLRR222 pKa = 11.84VRR224 pKa = 11.84LCDD227 pKa = 3.39NMNPLWRR234 pKa = 11.84ALAFWRR240 pKa = 11.84YY241 pKa = 9.67KK242 pKa = 10.74SLFMDD247 pKa = 4.0AKK249 pKa = 9.99FVNSGGLVFKK259 pKa = 10.8QKK261 pKa = 10.79FIGVMKK267 pKa = 9.94SGCVNTIITNSIAQILLHH285 pKa = 6.78LRR287 pKa = 11.84ICLEE291 pKa = 4.32LGLDD295 pKa = 3.52ISWIFSLGDD304 pKa = 3.33DD305 pKa = 4.12TLQEE309 pKa = 4.29LLSDD313 pKa = 3.74PRR315 pKa = 11.84YY316 pKa = 10.6LEE318 pKa = 4.05LLKK321 pKa = 10.77QFSLVKK327 pKa = 9.94EE328 pKa = 3.95CVIANEE334 pKa = 4.15FAGMRR339 pKa = 11.84YY340 pKa = 9.89AGGVVDD346 pKa = 4.78PLYY349 pKa = 10.61KK350 pKa = 10.42GKK352 pKa = 9.53HH353 pKa = 5.3AFNILHH359 pKa = 5.98VNPKK363 pKa = 9.7FGQEE367 pKa = 3.44IAEE370 pKa = 4.88GYY372 pKa = 9.71SILYY376 pKa = 9.12HH377 pKa = 6.33RR378 pKa = 11.84SPRR381 pKa = 11.84GSFIKK386 pKa = 10.54QIFKK390 pKa = 11.24DD391 pKa = 3.47MGYY394 pKa = 8.87TIPSDD399 pKa = 4.1DD400 pKa = 4.7ALDD403 pKa = 4.99LIWDD407 pKa = 3.94GEE409 pKa = 4.34EE410 pKa = 3.76
MM1 pKa = 7.61ASDD4 pKa = 4.02FTWPLLGGDD13 pKa = 3.71AEE15 pKa = 4.42LRR17 pKa = 11.84SLLTHH22 pKa = 5.46VQMSEE27 pKa = 3.76GLRR30 pKa = 11.84EE31 pKa = 4.77DD32 pKa = 4.58GPSHH36 pKa = 6.42QIKK39 pKa = 9.16TKK41 pKa = 10.58LLDD44 pKa = 3.3IAEE47 pKa = 4.84DD48 pKa = 3.58MFKK51 pKa = 10.16PARR54 pKa = 11.84WRR56 pKa = 11.84IPDD59 pKa = 3.93DD60 pKa = 3.57FLSFNHH66 pKa = 6.07YY67 pKa = 9.38MRR69 pKa = 11.84TLNQLVWTSSPGLPYY84 pKa = 9.93MRR86 pKa = 11.84TYY88 pKa = 10.39PNNGGMFGVVDD99 pKa = 4.28GVPNPEE105 pKa = 3.88KK106 pKa = 10.5CRR108 pKa = 11.84YY109 pKa = 6.63WWTIVQQKK117 pKa = 9.68IRR119 pKa = 11.84DD120 pKa = 3.67QFSDD124 pKa = 3.82YY125 pKa = 10.71IRR127 pKa = 11.84VFIKK131 pKa = 10.33QEE133 pKa = 4.12PITRR137 pKa = 11.84KK138 pKa = 9.98KK139 pKa = 10.57FEE141 pKa = 4.07QNRR144 pKa = 11.84FRR146 pKa = 11.84LIFSVSVLDD155 pKa = 4.56QIIDD159 pKa = 3.35HH160 pKa = 6.2MLFGDD165 pKa = 4.09FNDD168 pKa = 3.94LVIEE172 pKa = 3.99NWYY175 pKa = 10.19NLPTKK180 pKa = 10.53VGWSPLGGGWKK191 pKa = 9.93IIPKK195 pKa = 9.31SKK197 pKa = 10.2CHH199 pKa = 6.93AIDD202 pKa = 3.1KK203 pKa = 10.08RR204 pKa = 11.84AWDD207 pKa = 3.59WTVKK211 pKa = 8.5MWLIEE216 pKa = 4.19LEE218 pKa = 3.87FMLRR222 pKa = 11.84VRR224 pKa = 11.84LCDD227 pKa = 3.39NMNPLWRR234 pKa = 11.84ALAFWRR240 pKa = 11.84YY241 pKa = 9.67KK242 pKa = 10.74SLFMDD247 pKa = 4.0AKK249 pKa = 9.99FVNSGGLVFKK259 pKa = 10.8QKK261 pKa = 10.79FIGVMKK267 pKa = 9.94SGCVNTIITNSIAQILLHH285 pKa = 6.78LRR287 pKa = 11.84ICLEE291 pKa = 4.32LGLDD295 pKa = 3.52ISWIFSLGDD304 pKa = 3.33DD305 pKa = 4.12TLQEE309 pKa = 4.29LLSDD313 pKa = 3.74PRR315 pKa = 11.84YY316 pKa = 10.6LEE318 pKa = 4.05LLKK321 pKa = 10.77QFSLVKK327 pKa = 9.94EE328 pKa = 3.95CVIANEE334 pKa = 4.15FAGMRR339 pKa = 11.84YY340 pKa = 9.89AGGVVDD346 pKa = 4.78PLYY349 pKa = 10.61KK350 pKa = 10.42GKK352 pKa = 9.53HH353 pKa = 5.3AFNILHH359 pKa = 5.98VNPKK363 pKa = 9.7FGQEE367 pKa = 3.44IAEE370 pKa = 4.88GYY372 pKa = 9.71SILYY376 pKa = 9.12HH377 pKa = 6.33RR378 pKa = 11.84SPRR381 pKa = 11.84GSFIKK386 pKa = 10.54QIFKK390 pKa = 11.24DD391 pKa = 3.47MGYY394 pKa = 8.87TIPSDD399 pKa = 4.1DD400 pKa = 4.7ALDD403 pKa = 4.99LIWDD407 pKa = 3.94GEE409 pKa = 4.34EE410 pKa = 3.76
Molecular weight: 47.64 kDa
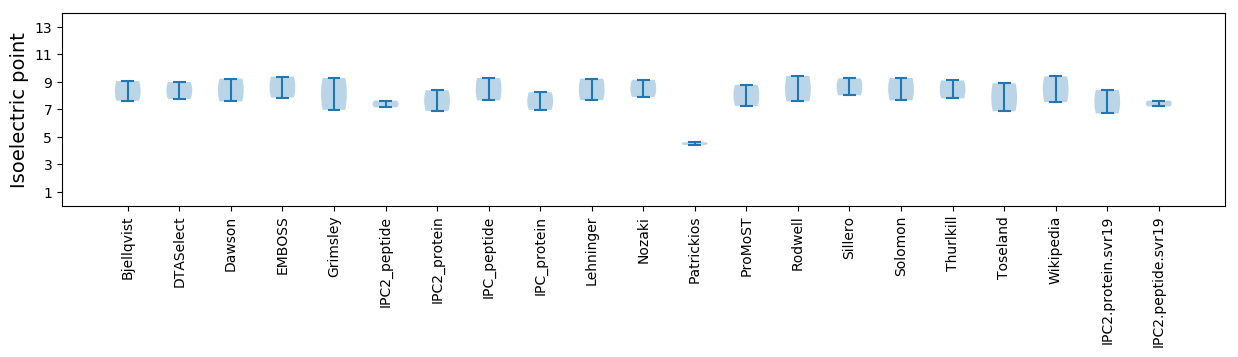
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEQ3|A0A1L3KEQ3_9VIRU C2H2-type domain-containing protein OS=Hubei sobemo-like virus 23 OX=1923209 PE=4 SV=1
MM1 pKa = 7.78FITEE5 pKa = 4.75FVSKK9 pKa = 10.3ILSNIRR15 pKa = 11.84NAIVPFVMAYY25 pKa = 8.98ILYY28 pKa = 9.6YY29 pKa = 10.89FMDD32 pKa = 3.96RR33 pKa = 11.84KK34 pKa = 10.46EE35 pKa = 4.32EE36 pKa = 4.24YY37 pKa = 9.79IDD39 pKa = 3.72TYY41 pKa = 11.45DD42 pKa = 3.72AVIDD46 pKa = 3.92SVRR49 pKa = 11.84PKK51 pKa = 10.49IVVQPSAEE59 pKa = 3.82FDD61 pKa = 3.7DD62 pKa = 6.37FIDD65 pKa = 4.12EE66 pKa = 4.55FLSKK70 pKa = 10.58AWTHH74 pKa = 4.38IRR76 pKa = 11.84RR77 pKa = 11.84YY78 pKa = 10.25VSLKK82 pKa = 9.13GHH84 pKa = 6.99RR85 pKa = 11.84IWYY88 pKa = 9.12LIVKK92 pKa = 9.06VLLVVLTVYY101 pKa = 11.06LLYY104 pKa = 10.8KK105 pKa = 10.25GIRR108 pKa = 11.84KK109 pKa = 8.94LFSFIKK115 pKa = 9.8IKK117 pKa = 10.2MYY119 pKa = 10.41DD120 pKa = 3.25FCGVKK125 pKa = 10.2DD126 pKa = 4.12FRR128 pKa = 11.84GEE130 pKa = 3.7AMKK133 pKa = 10.71AGSSFGKK140 pKa = 9.03GTVPDD145 pKa = 3.97YY146 pKa = 10.59QVEE149 pKa = 4.11IHH151 pKa = 7.02RR152 pKa = 11.84SGTFMFSFSGYY163 pKa = 7.41GLRR166 pKa = 11.84YY167 pKa = 9.59GDD169 pKa = 3.7VLVVPTHH176 pKa = 6.24VIGNSRR182 pKa = 11.84DD183 pKa = 3.39FKK185 pKa = 11.4LVGKK189 pKa = 8.26KK190 pKa = 7.3TAYY193 pKa = 9.59ISLDD197 pKa = 3.64FVASTLMNDD206 pKa = 3.03VSYY209 pKa = 11.49VRR211 pKa = 11.84LGTDD215 pKa = 2.49VWSALGTKK223 pKa = 8.34SCKK226 pKa = 9.96YY227 pKa = 9.82IKK229 pKa = 10.08PNYY232 pKa = 7.78TPQVVTIVGKK242 pKa = 10.47QGATSGILRR251 pKa = 11.84KK252 pKa = 9.39SEE254 pKa = 4.15CEE256 pKa = 3.78FMMEE260 pKa = 4.42YY261 pKa = 10.16EE262 pKa = 4.57GSTIPGMSGAAYY274 pKa = 7.62FTQAGPLAMHH284 pKa = 6.72SGVVLGRR291 pKa = 11.84TNVGYY296 pKa = 8.64TINAICMEE304 pKa = 4.43VDD306 pKa = 3.28TTFPPSVVGEE316 pKa = 4.01GRR318 pKa = 11.84IADD321 pKa = 3.66ANEE324 pKa = 4.23DD325 pKa = 3.95YY326 pKa = 10.9EE327 pKa = 4.68FSGRR331 pKa = 11.84RR332 pKa = 11.84KK333 pKa = 8.68KK334 pKa = 10.64AAWRR338 pKa = 11.84EE339 pKa = 4.24SKK341 pKa = 10.88VKK343 pKa = 10.82SSWDD347 pKa = 3.27NSSIKK352 pKa = 10.48ARR354 pKa = 11.84LGFADD359 pKa = 3.85VSDD362 pKa = 3.68EE363 pKa = 4.66RR364 pKa = 11.84IAEE367 pKa = 4.19LEE369 pKa = 4.05RR370 pKa = 11.84FANADD375 pKa = 3.85DD376 pKa = 4.31AWTKK380 pKa = 10.61DD381 pKa = 2.88MKK383 pKa = 10.25MDD385 pKa = 3.89YY386 pKa = 10.17NVKK389 pKa = 10.66LNFEE393 pKa = 4.69GEE395 pKa = 4.13NFDD398 pKa = 3.7PSKK401 pKa = 11.03KK402 pKa = 9.96IFATDD407 pKa = 3.0GHH409 pKa = 5.79GKK411 pKa = 9.8KK412 pKa = 10.57VIILGTSDD420 pKa = 4.42LNFSGFKK427 pKa = 9.9PYY429 pKa = 10.74SATASGQAPDD439 pKa = 4.0EE440 pKa = 4.05PQIRR444 pKa = 11.84IGAVEE449 pKa = 4.33PSLGDD454 pKa = 2.97IVRR457 pKa = 11.84DD458 pKa = 3.38HH459 pKa = 6.69EE460 pKa = 4.26RR461 pKa = 11.84RR462 pKa = 11.84IVALEE467 pKa = 3.83AKK469 pKa = 9.99IKK471 pKa = 10.31SLQSKK476 pKa = 9.29PPGYY480 pKa = 10.87VCGHH484 pKa = 5.92CAKK487 pKa = 10.38PFDD490 pKa = 4.48LEE492 pKa = 4.07EE493 pKa = 5.34SLMQHH498 pKa = 6.02VKK500 pKa = 10.21SKK502 pKa = 10.36HH503 pKa = 5.29IKK505 pKa = 10.35GEE507 pKa = 4.07SAYY510 pKa = 9.65KK511 pKa = 10.6DD512 pKa = 3.51HH513 pKa = 7.73FDD515 pKa = 6.35DD516 pKa = 5.78ILNKK520 pKa = 10.26DD521 pKa = 3.13KK522 pKa = 10.64KK523 pKa = 9.33TFLEE527 pKa = 4.3QKK529 pKa = 10.27PSPTKK534 pKa = 9.88NAPNSRR540 pKa = 11.84STSRR544 pKa = 11.84SKK546 pKa = 10.68EE547 pKa = 3.73KK548 pKa = 10.46KK549 pKa = 9.06VPSTSRR555 pKa = 11.84EE556 pKa = 4.06GNRR559 pKa = 11.84SS560 pKa = 3.13
MM1 pKa = 7.78FITEE5 pKa = 4.75FVSKK9 pKa = 10.3ILSNIRR15 pKa = 11.84NAIVPFVMAYY25 pKa = 8.98ILYY28 pKa = 9.6YY29 pKa = 10.89FMDD32 pKa = 3.96RR33 pKa = 11.84KK34 pKa = 10.46EE35 pKa = 4.32EE36 pKa = 4.24YY37 pKa = 9.79IDD39 pKa = 3.72TYY41 pKa = 11.45DD42 pKa = 3.72AVIDD46 pKa = 3.92SVRR49 pKa = 11.84PKK51 pKa = 10.49IVVQPSAEE59 pKa = 3.82FDD61 pKa = 3.7DD62 pKa = 6.37FIDD65 pKa = 4.12EE66 pKa = 4.55FLSKK70 pKa = 10.58AWTHH74 pKa = 4.38IRR76 pKa = 11.84RR77 pKa = 11.84YY78 pKa = 10.25VSLKK82 pKa = 9.13GHH84 pKa = 6.99RR85 pKa = 11.84IWYY88 pKa = 9.12LIVKK92 pKa = 9.06VLLVVLTVYY101 pKa = 11.06LLYY104 pKa = 10.8KK105 pKa = 10.25GIRR108 pKa = 11.84KK109 pKa = 8.94LFSFIKK115 pKa = 9.8IKK117 pKa = 10.2MYY119 pKa = 10.41DD120 pKa = 3.25FCGVKK125 pKa = 10.2DD126 pKa = 4.12FRR128 pKa = 11.84GEE130 pKa = 3.7AMKK133 pKa = 10.71AGSSFGKK140 pKa = 9.03GTVPDD145 pKa = 3.97YY146 pKa = 10.59QVEE149 pKa = 4.11IHH151 pKa = 7.02RR152 pKa = 11.84SGTFMFSFSGYY163 pKa = 7.41GLRR166 pKa = 11.84YY167 pKa = 9.59GDD169 pKa = 3.7VLVVPTHH176 pKa = 6.24VIGNSRR182 pKa = 11.84DD183 pKa = 3.39FKK185 pKa = 11.4LVGKK189 pKa = 8.26KK190 pKa = 7.3TAYY193 pKa = 9.59ISLDD197 pKa = 3.64FVASTLMNDD206 pKa = 3.03VSYY209 pKa = 11.49VRR211 pKa = 11.84LGTDD215 pKa = 2.49VWSALGTKK223 pKa = 8.34SCKK226 pKa = 9.96YY227 pKa = 9.82IKK229 pKa = 10.08PNYY232 pKa = 7.78TPQVVTIVGKK242 pKa = 10.47QGATSGILRR251 pKa = 11.84KK252 pKa = 9.39SEE254 pKa = 4.15CEE256 pKa = 3.78FMMEE260 pKa = 4.42YY261 pKa = 10.16EE262 pKa = 4.57GSTIPGMSGAAYY274 pKa = 7.62FTQAGPLAMHH284 pKa = 6.72SGVVLGRR291 pKa = 11.84TNVGYY296 pKa = 8.64TINAICMEE304 pKa = 4.43VDD306 pKa = 3.28TTFPPSVVGEE316 pKa = 4.01GRR318 pKa = 11.84IADD321 pKa = 3.66ANEE324 pKa = 4.23DD325 pKa = 3.95YY326 pKa = 10.9EE327 pKa = 4.68FSGRR331 pKa = 11.84RR332 pKa = 11.84KK333 pKa = 8.68KK334 pKa = 10.64AAWRR338 pKa = 11.84EE339 pKa = 4.24SKK341 pKa = 10.88VKK343 pKa = 10.82SSWDD347 pKa = 3.27NSSIKK352 pKa = 10.48ARR354 pKa = 11.84LGFADD359 pKa = 3.85VSDD362 pKa = 3.68EE363 pKa = 4.66RR364 pKa = 11.84IAEE367 pKa = 4.19LEE369 pKa = 4.05RR370 pKa = 11.84FANADD375 pKa = 3.85DD376 pKa = 4.31AWTKK380 pKa = 10.61DD381 pKa = 2.88MKK383 pKa = 10.25MDD385 pKa = 3.89YY386 pKa = 10.17NVKK389 pKa = 10.66LNFEE393 pKa = 4.69GEE395 pKa = 4.13NFDD398 pKa = 3.7PSKK401 pKa = 11.03KK402 pKa = 9.96IFATDD407 pKa = 3.0GHH409 pKa = 5.79GKK411 pKa = 9.8KK412 pKa = 10.57VIILGTSDD420 pKa = 4.42LNFSGFKK427 pKa = 9.9PYY429 pKa = 10.74SATASGQAPDD439 pKa = 4.0EE440 pKa = 4.05PQIRR444 pKa = 11.84IGAVEE449 pKa = 4.33PSLGDD454 pKa = 2.97IVRR457 pKa = 11.84DD458 pKa = 3.38HH459 pKa = 6.69EE460 pKa = 4.26RR461 pKa = 11.84RR462 pKa = 11.84IVALEE467 pKa = 3.83AKK469 pKa = 9.99IKK471 pKa = 10.31SLQSKK476 pKa = 9.29PPGYY480 pKa = 10.87VCGHH484 pKa = 5.92CAKK487 pKa = 10.38PFDD490 pKa = 4.48LEE492 pKa = 4.07EE493 pKa = 5.34SLMQHH498 pKa = 6.02VKK500 pKa = 10.21SKK502 pKa = 10.36HH503 pKa = 5.29IKK505 pKa = 10.35GEE507 pKa = 4.07SAYY510 pKa = 9.65KK511 pKa = 10.6DD512 pKa = 3.51HH513 pKa = 7.73FDD515 pKa = 6.35DD516 pKa = 5.78ILNKK520 pKa = 10.26DD521 pKa = 3.13KK522 pKa = 10.64KK523 pKa = 9.33TFLEE527 pKa = 4.3QKK529 pKa = 10.27PSPTKK534 pKa = 9.88NAPNSRR540 pKa = 11.84STSRR544 pKa = 11.84SKK546 pKa = 10.68EE547 pKa = 3.73KK548 pKa = 10.46KK549 pKa = 9.06VPSTSRR555 pKa = 11.84EE556 pKa = 4.06GNRR559 pKa = 11.84SS560 pKa = 3.13
Molecular weight: 62.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
970 |
410 |
560 |
485.0 |
55.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.361 ± 0.909 | 1.237 ± 0.141 |
6.598 ± 0.144 | 5.155 ± 0.324 |
5.773 ± 0.202 | 7.216 ± 0.089 |
2.062 ± 0.083 | 7.113 ± 0.431 |
7.835 ± 0.93 | 8.041 ± 1.828 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.99 ± 0.265 | 3.608 ± 0.335 |
4.33 ± 0.19 | 2.577 ± 0.674 |
5.258 ± 0.067 | 7.629 ± 1.106 |
4.33 ± 0.57 | 6.804 ± 0.896 |
2.165 ± 0.93 | 3.918 ± 0.465 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
