
Pseudoxanthomonas sp. GM95
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Pseudoxanthomonas; unclassified Pseudoxanthomonas
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
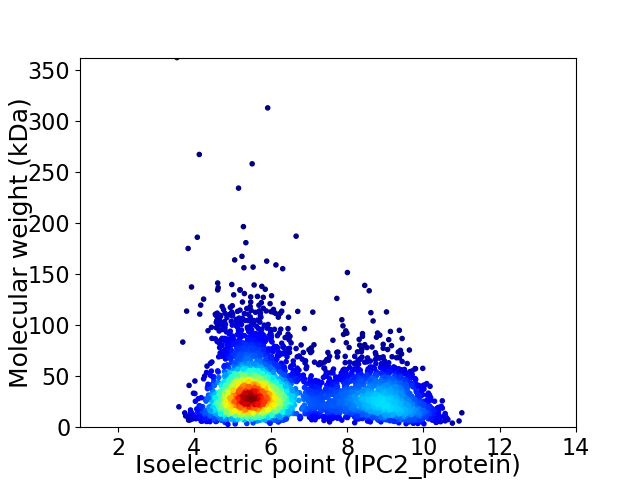
Virtual 2D-PAGE plot for 4071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7XR37|A0A1H7XR37_9GAMM D-galactonate transporter OS=Pseudoxanthomonas sp. GM95 OX=1881043 GN=SAMN05428989_3636 PE=4 SV=1
MM1 pKa = 7.68PSSVMPSARR10 pKa = 11.84RR11 pKa = 11.84GRR13 pKa = 11.84TSSHH17 pKa = 6.97RR18 pKa = 11.84ILPALSCNPLTAALLLVLALPAAATAPASGNAAAAPATQASATGTATTQDD68 pKa = 3.95DD69 pKa = 4.59DD70 pKa = 4.22TTSVQPQSAEE80 pKa = 3.56AAASYY85 pKa = 9.34FAANGLGDD93 pKa = 4.02GSDD96 pKa = 3.96DD97 pKa = 3.6ATASGTNAVAVGAGADD113 pKa = 3.37ATGDD117 pKa = 3.5YY118 pKa = 10.66GVAIGSHH125 pKa = 5.24ATVTDD130 pKa = 3.97AYY132 pKa = 10.55GIAIGHH138 pKa = 6.21NVTAGNTAIAMGDD151 pKa = 3.54GAQATGQSAVAIGGSFFGSTYY172 pKa = 10.58GDD174 pKa = 3.15SDD176 pKa = 3.68GARR179 pKa = 11.84ATGFGAVSLGAYY191 pKa = 9.9AGATGSTSTALGWSSSAIEE210 pKa = 4.31SGTVAVGSSAHH221 pKa = 6.39AEE223 pKa = 4.17GVYY226 pKa = 10.32AAAFGSSTYY235 pKa = 10.13ATGTASTALGYY246 pKa = 8.64GTVASNDD253 pKa = 3.07ATVAIGTFGQADD265 pKa = 4.3GFAGVAVGYY274 pKa = 9.71YY275 pKa = 8.93ATASGDD281 pKa = 3.29GATALGEE288 pKa = 4.16SALAQGQQSVAIGASNAGVYY308 pKa = 7.82TQANGLGSVTVGAGSWALSDD328 pKa = 3.76YY329 pKa = 9.84GVAMGFDD336 pKa = 3.65SHH338 pKa = 8.07ADD340 pKa = 3.2AMYY343 pKa = 8.82ATALGAQAISTSTSATALGAQSFSDD368 pKa = 3.72GDD370 pKa = 3.44EE371 pKa = 4.09ATAVGYY377 pKa = 9.73IASASGLGASAFGAGASAFGDD398 pKa = 3.47GGLALGTNSAVIGANSVALGAGSFADD424 pKa = 3.77RR425 pKa = 11.84DD426 pKa = 3.84NTVSVGVAGAEE437 pKa = 3.65RR438 pKa = 11.84QVSNVAAGTEE448 pKa = 4.06DD449 pKa = 3.45TDD451 pKa = 4.0AVNVAQLKK459 pKa = 11.07SMTQNIGDD467 pKa = 4.15DD468 pKa = 3.84DD469 pKa = 6.02LYY471 pKa = 11.4FSANGAGDD479 pKa = 3.85GTDD482 pKa = 2.96TATASGTGSLAAGSSAEE499 pKa = 3.81ATGNYY504 pKa = 8.77GVALGSRR511 pKa = 11.84AVVSDD516 pKa = 4.14AYY518 pKa = 10.7GIAIGHH524 pKa = 5.85NVQAGNTAIAMGDD537 pKa = 3.54GAQATGQSAVAIGGSYY553 pKa = 10.18FGSAFGDD560 pKa = 3.69SIGAIGSGFGAVALGAAASATGSTSTAVGWAAHH593 pKa = 6.0TDD595 pKa = 4.02FSGGLAMGASAGAMADD611 pKa = 3.52GAVALGASSYY621 pKa = 8.06VTGIQGTGVGYY632 pKa = 9.88GAYY635 pKa = 8.7VTGTVGVALGGYY647 pKa = 8.78SEE649 pKa = 4.41VTGAFAMALGYY660 pKa = 9.87GAQTVGDD667 pKa = 3.65GGIAVGEE674 pKa = 3.97AALAQGQQSVAIGATNGGISTQANGLGSTTVGAGSWALSDD714 pKa = 3.67YY715 pKa = 10.87GVALGFDD722 pKa = 3.69SHH724 pKa = 8.08ADD726 pKa = 3.34AMYY729 pKa = 9.26STALGAQALSTSTSATALGASTFADD754 pKa = 3.61GDD756 pKa = 3.59EE757 pKa = 4.26ATAVGYY763 pKa = 9.67IASASGVGAAAFGAGASAFGDD784 pKa = 3.38GGLAVGYY791 pKa = 9.5NAAAVGLNSVALGAGSYY808 pKa = 10.98ADD810 pKa = 4.26RR811 pKa = 11.84DD812 pKa = 3.71NSVSFGVAGAEE823 pKa = 3.65RR824 pKa = 11.84QLTNIAAGTEE834 pKa = 4.01DD835 pKa = 3.64TDD837 pKa = 4.88AVNLSQLHH845 pKa = 6.15ALADD849 pKa = 3.9TLILSGGATVDD860 pKa = 3.9GMGEE864 pKa = 3.64MAAAFGGGAGYY875 pKa = 10.05SAAGGFVLPSYY886 pKa = 10.29SIQGGSYY893 pKa = 10.05TNVGDD898 pKa = 5.04ALTALDD904 pKa = 4.35GAFSGLSDD912 pKa = 3.83RR913 pKa = 11.84LASLEE918 pKa = 4.09TAPAAGSGAGGVAVGGGAADD938 pKa = 4.0GASVADD944 pKa = 4.22GSNGVAVGAGAQTGGEE960 pKa = 4.13NGTAVGGGAYY970 pKa = 10.11AAGPNDD976 pKa = 3.58TAIGGNAKK984 pKa = 10.01VNADD988 pKa = 2.9GSTAVGANTAIAAEE1002 pKa = 4.2ATNAVAVGEE1011 pKa = 4.35SASVSVASGTAVGQGASVTAEE1032 pKa = 3.88GAVALGQGSVADD1044 pKa = 4.17RR1045 pKa = 11.84ANTVSVGSTGSEE1057 pKa = 3.56RR1058 pKa = 11.84QVTHH1062 pKa = 6.01VAAGTAATDD1071 pKa = 3.62AANVGQMQAGDD1082 pKa = 3.89AQSVATANAYY1092 pKa = 8.63TDD1094 pKa = 3.54TTATRR1099 pKa = 11.84TLTSANSYY1107 pKa = 9.7TDD1109 pKa = 3.4QKK1111 pKa = 9.15LTAMDD1116 pKa = 4.3DD1117 pKa = 3.74RR1118 pKa = 11.84FTQLSNDD1125 pKa = 3.72VGHH1128 pKa = 7.35KK1129 pKa = 10.0LAAQDD1134 pKa = 3.92KK1135 pKa = 9.9RR1136 pKa = 11.84IDD1138 pKa = 3.35RR1139 pKa = 11.84MGAMGSAMMNMAINAANSKK1158 pKa = 8.55SAKK1161 pKa = 9.67GRR1163 pKa = 11.84IAAGAGWQNGEE1174 pKa = 3.93AALSVGYY1181 pKa = 10.28AKK1183 pKa = 10.6QIGEE1187 pKa = 4.15RR1188 pKa = 11.84ASFSIGGAFSGDD1200 pKa = 3.51DD1201 pKa = 3.3TSAGVGFGIDD1211 pKa = 3.25LL1212 pKa = 4.11
MM1 pKa = 7.68PSSVMPSARR10 pKa = 11.84RR11 pKa = 11.84GRR13 pKa = 11.84TSSHH17 pKa = 6.97RR18 pKa = 11.84ILPALSCNPLTAALLLVLALPAAATAPASGNAAAAPATQASATGTATTQDD68 pKa = 3.95DD69 pKa = 4.59DD70 pKa = 4.22TTSVQPQSAEE80 pKa = 3.56AAASYY85 pKa = 9.34FAANGLGDD93 pKa = 4.02GSDD96 pKa = 3.96DD97 pKa = 3.6ATASGTNAVAVGAGADD113 pKa = 3.37ATGDD117 pKa = 3.5YY118 pKa = 10.66GVAIGSHH125 pKa = 5.24ATVTDD130 pKa = 3.97AYY132 pKa = 10.55GIAIGHH138 pKa = 6.21NVTAGNTAIAMGDD151 pKa = 3.54GAQATGQSAVAIGGSFFGSTYY172 pKa = 10.58GDD174 pKa = 3.15SDD176 pKa = 3.68GARR179 pKa = 11.84ATGFGAVSLGAYY191 pKa = 9.9AGATGSTSTALGWSSSAIEE210 pKa = 4.31SGTVAVGSSAHH221 pKa = 6.39AEE223 pKa = 4.17GVYY226 pKa = 10.32AAAFGSSTYY235 pKa = 10.13ATGTASTALGYY246 pKa = 8.64GTVASNDD253 pKa = 3.07ATVAIGTFGQADD265 pKa = 4.3GFAGVAVGYY274 pKa = 9.71YY275 pKa = 8.93ATASGDD281 pKa = 3.29GATALGEE288 pKa = 4.16SALAQGQQSVAIGASNAGVYY308 pKa = 7.82TQANGLGSVTVGAGSWALSDD328 pKa = 3.76YY329 pKa = 9.84GVAMGFDD336 pKa = 3.65SHH338 pKa = 8.07ADD340 pKa = 3.2AMYY343 pKa = 8.82ATALGAQAISTSTSATALGAQSFSDD368 pKa = 3.72GDD370 pKa = 3.44EE371 pKa = 4.09ATAVGYY377 pKa = 9.73IASASGLGASAFGAGASAFGDD398 pKa = 3.47GGLALGTNSAVIGANSVALGAGSFADD424 pKa = 3.77RR425 pKa = 11.84DD426 pKa = 3.84NTVSVGVAGAEE437 pKa = 3.65RR438 pKa = 11.84QVSNVAAGTEE448 pKa = 4.06DD449 pKa = 3.45TDD451 pKa = 4.0AVNVAQLKK459 pKa = 11.07SMTQNIGDD467 pKa = 4.15DD468 pKa = 3.84DD469 pKa = 6.02LYY471 pKa = 11.4FSANGAGDD479 pKa = 3.85GTDD482 pKa = 2.96TATASGTGSLAAGSSAEE499 pKa = 3.81ATGNYY504 pKa = 8.77GVALGSRR511 pKa = 11.84AVVSDD516 pKa = 4.14AYY518 pKa = 10.7GIAIGHH524 pKa = 5.85NVQAGNTAIAMGDD537 pKa = 3.54GAQATGQSAVAIGGSYY553 pKa = 10.18FGSAFGDD560 pKa = 3.69SIGAIGSGFGAVALGAAASATGSTSTAVGWAAHH593 pKa = 6.0TDD595 pKa = 4.02FSGGLAMGASAGAMADD611 pKa = 3.52GAVALGASSYY621 pKa = 8.06VTGIQGTGVGYY632 pKa = 9.88GAYY635 pKa = 8.7VTGTVGVALGGYY647 pKa = 8.78SEE649 pKa = 4.41VTGAFAMALGYY660 pKa = 9.87GAQTVGDD667 pKa = 3.65GGIAVGEE674 pKa = 3.97AALAQGQQSVAIGATNGGISTQANGLGSTTVGAGSWALSDD714 pKa = 3.67YY715 pKa = 10.87GVALGFDD722 pKa = 3.69SHH724 pKa = 8.08ADD726 pKa = 3.34AMYY729 pKa = 9.26STALGAQALSTSTSATALGASTFADD754 pKa = 3.61GDD756 pKa = 3.59EE757 pKa = 4.26ATAVGYY763 pKa = 9.67IASASGVGAAAFGAGASAFGDD784 pKa = 3.38GGLAVGYY791 pKa = 9.5NAAAVGLNSVALGAGSYY808 pKa = 10.98ADD810 pKa = 4.26RR811 pKa = 11.84DD812 pKa = 3.71NSVSFGVAGAEE823 pKa = 3.65RR824 pKa = 11.84QLTNIAAGTEE834 pKa = 4.01DD835 pKa = 3.64TDD837 pKa = 4.88AVNLSQLHH845 pKa = 6.15ALADD849 pKa = 3.9TLILSGGATVDD860 pKa = 3.9GMGEE864 pKa = 3.64MAAAFGGGAGYY875 pKa = 10.05SAAGGFVLPSYY886 pKa = 10.29SIQGGSYY893 pKa = 10.05TNVGDD898 pKa = 5.04ALTALDD904 pKa = 4.35GAFSGLSDD912 pKa = 3.83RR913 pKa = 11.84LASLEE918 pKa = 4.09TAPAAGSGAGGVAVGGGAADD938 pKa = 4.0GASVADD944 pKa = 4.22GSNGVAVGAGAQTGGEE960 pKa = 4.13NGTAVGGGAYY970 pKa = 10.11AAGPNDD976 pKa = 3.58TAIGGNAKK984 pKa = 10.01VNADD988 pKa = 2.9GSTAVGANTAIAAEE1002 pKa = 4.2ATNAVAVGEE1011 pKa = 4.35SASVSVASGTAVGQGASVTAEE1032 pKa = 3.88GAVALGQGSVADD1044 pKa = 4.17RR1045 pKa = 11.84ANTVSVGSTGSEE1057 pKa = 3.56RR1058 pKa = 11.84QVTHH1062 pKa = 6.01VAAGTAATDD1071 pKa = 3.62AANVGQMQAGDD1082 pKa = 3.89AQSVATANAYY1092 pKa = 8.63TDD1094 pKa = 3.54TTATRR1099 pKa = 11.84TLTSANSYY1107 pKa = 9.7TDD1109 pKa = 3.4QKK1111 pKa = 9.15LTAMDD1116 pKa = 4.3DD1117 pKa = 3.74RR1118 pKa = 11.84FTQLSNDD1125 pKa = 3.72VGHH1128 pKa = 7.35KK1129 pKa = 10.0LAAQDD1134 pKa = 3.92KK1135 pKa = 9.9RR1136 pKa = 11.84IDD1138 pKa = 3.35RR1139 pKa = 11.84MGAMGSAMMNMAINAANSKK1158 pKa = 8.55SAKK1161 pKa = 9.67GRR1163 pKa = 11.84IAAGAGWQNGEE1174 pKa = 3.93AALSVGYY1181 pKa = 10.28AKK1183 pKa = 10.6QIGEE1187 pKa = 4.15RR1188 pKa = 11.84ASFSIGGAFSGDD1200 pKa = 3.51DD1201 pKa = 3.3TSAGVGFGIDD1211 pKa = 3.25LL1212 pKa = 4.11
Molecular weight: 113.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7U7P7|A0A1H7U7P7_9GAMM GTPase Der OS=Pseudoxanthomonas sp. GM95 OX=1881043 GN=der PE=3 SV=1
MM1 pKa = 8.08ASTTQLKK8 pKa = 10.19IGAGALWIAGLATLVAALLLRR29 pKa = 11.84QPLLLPVGAVLIGVGRR45 pKa = 11.84TAMTRR50 pKa = 11.84ILRR53 pKa = 11.84RR54 pKa = 11.84TLPRR58 pKa = 4.93
MM1 pKa = 8.08ASTTQLKK8 pKa = 10.19IGAGALWIAGLATLVAALLLRR29 pKa = 11.84QPLLLPVGAVLIGVGRR45 pKa = 11.84TAMTRR50 pKa = 11.84ILRR53 pKa = 11.84RR54 pKa = 11.84TLPRR58 pKa = 4.93
Molecular weight: 6.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1401671 |
29 |
3738 |
344.3 |
37.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.056 ± 0.061 | 0.794 ± 0.01 |
5.884 ± 0.034 | 4.84 ± 0.033 |
3.382 ± 0.025 | 8.579 ± 0.035 |
2.207 ± 0.02 | 4.112 ± 0.027 |
2.918 ± 0.026 | 10.796 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.193 ± 0.018 | 2.566 ± 0.032 |
5.338 ± 0.03 | 4.103 ± 0.025 |
6.848 ± 0.043 | 5.444 ± 0.031 |
5.435 ± 0.039 | 7.524 ± 0.036 |
1.561 ± 0.018 | 2.422 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
