
Eschrichtius robustus (California gray whale) (Eschrichtius gibbosus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria;
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
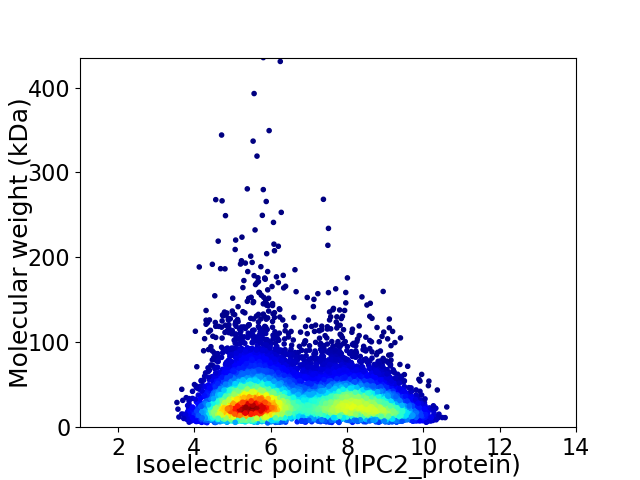
Virtual 2D-PAGE plot for 10855 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

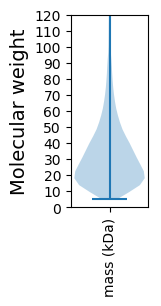
Protein with the lowest isoelectric point:
>tr|A0A2F0AY62|A0A2F0AY62_ESCRO GDP-fucose transporter 1 (Fragment) OS=Eschrichtius robustus OX=9764 GN=ESR_30128 PE=3 SV=1
MM1 pKa = 7.9DD2 pKa = 4.03HH3 pKa = 6.71TAFPCGCCRR12 pKa = 11.84EE13 pKa = 4.32GCEE16 pKa = 4.09NPKK19 pKa = 10.53GRR21 pKa = 11.84VEE23 pKa = 4.13FNQARR28 pKa = 11.84VQTHH32 pKa = 7.12FIHH35 pKa = 6.13TLTRR39 pKa = 11.84LQLEE43 pKa = 4.23QGAEE47 pKa = 4.07SLGEE51 pKa = 4.09LEE53 pKa = 5.72APAQGATFSPNEE65 pKa = 3.96QVLAPTFPLAKK76 pKa = 9.66PPVSSEE82 pKa = 4.27LGDD85 pKa = 3.79NSCSSDD91 pKa = 3.31MTDD94 pKa = 2.81SSTASSSSEE103 pKa = 3.9APSGSAHH110 pKa = 6.96PALPGPSFQPGVDD123 pKa = 3.76DD124 pKa = 5.97DD125 pKa = 4.2SLARR129 pKa = 11.84ILSFSDD135 pKa = 3.16SDD137 pKa = 3.74LGGDD141 pKa = 3.78GEE143 pKa = 4.36EE144 pKa = 4.38EE145 pKa = 4.26EE146 pKa = 4.39EE147 pKa = 4.64GGVGSLDD154 pKa = 3.42NLSCFHH160 pKa = 7.14PADD163 pKa = 3.3IFGTGDD169 pKa = 3.56PAGLASWTHH178 pKa = 6.1SYY180 pKa = 10.94SSSSLASGILDD191 pKa = 3.36EE192 pKa = 5.03NANLDD197 pKa = 3.57ASCFLNSGLEE207 pKa = 4.07SLGEE211 pKa = 4.1GSLPGPPVPAITDD224 pKa = 3.9ASQSSSVDD232 pKa = 3.62LSLSSCDD239 pKa = 3.95SFKK242 pKa = 11.06LLQALPDD249 pKa = 3.89YY250 pKa = 11.07SLGPHH255 pKa = 5.27YY256 pKa = 9.71TSQKK260 pKa = 10.32VSDD263 pKa = 3.91SLDD266 pKa = 3.48NLEE269 pKa = 4.97APHH272 pKa = 6.36FALPAFSPPGDD283 pKa = 3.41ASTCFLEE290 pKa = 5.53SIMGLPEE297 pKa = 3.84PAAEE301 pKa = 4.02ALAPFMDD308 pKa = 4.17GQLFEE313 pKa = 4.63DD314 pKa = 4.29TAPASLVEE322 pKa = 4.21PVSVV326 pKa = 3.42
MM1 pKa = 7.9DD2 pKa = 4.03HH3 pKa = 6.71TAFPCGCCRR12 pKa = 11.84EE13 pKa = 4.32GCEE16 pKa = 4.09NPKK19 pKa = 10.53GRR21 pKa = 11.84VEE23 pKa = 4.13FNQARR28 pKa = 11.84VQTHH32 pKa = 7.12FIHH35 pKa = 6.13TLTRR39 pKa = 11.84LQLEE43 pKa = 4.23QGAEE47 pKa = 4.07SLGEE51 pKa = 4.09LEE53 pKa = 5.72APAQGATFSPNEE65 pKa = 3.96QVLAPTFPLAKK76 pKa = 9.66PPVSSEE82 pKa = 4.27LGDD85 pKa = 3.79NSCSSDD91 pKa = 3.31MTDD94 pKa = 2.81SSTASSSSEE103 pKa = 3.9APSGSAHH110 pKa = 6.96PALPGPSFQPGVDD123 pKa = 3.76DD124 pKa = 5.97DD125 pKa = 4.2SLARR129 pKa = 11.84ILSFSDD135 pKa = 3.16SDD137 pKa = 3.74LGGDD141 pKa = 3.78GEE143 pKa = 4.36EE144 pKa = 4.38EE145 pKa = 4.26EE146 pKa = 4.39EE147 pKa = 4.64GGVGSLDD154 pKa = 3.42NLSCFHH160 pKa = 7.14PADD163 pKa = 3.3IFGTGDD169 pKa = 3.56PAGLASWTHH178 pKa = 6.1SYY180 pKa = 10.94SSSSLASGILDD191 pKa = 3.36EE192 pKa = 5.03NANLDD197 pKa = 3.57ASCFLNSGLEE207 pKa = 4.07SLGEE211 pKa = 4.1GSLPGPPVPAITDD224 pKa = 3.9ASQSSSVDD232 pKa = 3.62LSLSSCDD239 pKa = 3.95SFKK242 pKa = 11.06LLQALPDD249 pKa = 3.89YY250 pKa = 11.07SLGPHH255 pKa = 5.27YY256 pKa = 9.71TSQKK260 pKa = 10.32VSDD263 pKa = 3.91SLDD266 pKa = 3.48NLEE269 pKa = 4.97APHH272 pKa = 6.36FALPAFSPPGDD283 pKa = 3.41ASTCFLEE290 pKa = 5.53SIMGLPEE297 pKa = 3.84PAAEE301 pKa = 4.02ALAPFMDD308 pKa = 4.17GQLFEE313 pKa = 4.63DD314 pKa = 4.29TAPASLVEE322 pKa = 4.21PVSVV326 pKa = 3.42
Molecular weight: 33.69 kDa
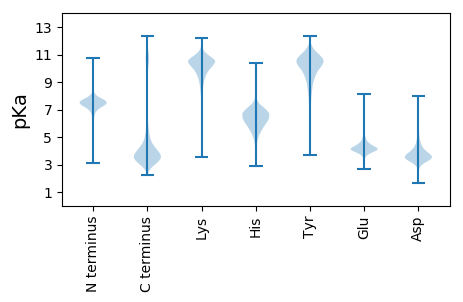
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2F0BK24|A0A2F0BK24_ESCRO 40S ribosomal protein S3 (Fragment) OS=Eschrichtius robustus OX=9764 GN=ESR_29911 PE=3 SV=1
MM1 pKa = 6.82MRR3 pKa = 11.84SSALLSSRR11 pKa = 11.84MPVQASSRR19 pKa = 11.84PWPRR23 pKa = 11.84ARR25 pKa = 11.84EE26 pKa = 3.92TEE28 pKa = 3.87RR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 4.11GCLQVCKK38 pKa = 10.74ARR40 pKa = 11.84GKK42 pKa = 10.1DD43 pKa = 3.34QSQRR47 pKa = 11.84PLRR50 pKa = 11.84RR51 pKa = 11.84PRR53 pKa = 11.84SPHH56 pKa = 5.61RR57 pKa = 11.84PRR59 pKa = 11.84WVYY62 pKa = 10.86QLTSVWMVFVVVASILTNGLVLAATMKK89 pKa = 10.23FKK91 pKa = 11.0KK92 pKa = 9.7LRR94 pKa = 11.84HH95 pKa = 5.28PLNWILVNLAIADD108 pKa = 3.97LAEE111 pKa = 4.34TIIAGTISIVNQMYY125 pKa = 9.65GYY127 pKa = 10.28FVLGHH132 pKa = 6.48PLCIVEE138 pKa = 5.28GYY140 pKa = 7.23TVCLWGVIEE149 pKa = 4.02AHH151 pKa = 6.37LL152 pKa = 4.08
MM1 pKa = 6.82MRR3 pKa = 11.84SSALLSSRR11 pKa = 11.84MPVQASSRR19 pKa = 11.84PWPRR23 pKa = 11.84ARR25 pKa = 11.84EE26 pKa = 3.92TEE28 pKa = 3.87RR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 4.11GCLQVCKK38 pKa = 10.74ARR40 pKa = 11.84GKK42 pKa = 10.1DD43 pKa = 3.34QSQRR47 pKa = 11.84PLRR50 pKa = 11.84RR51 pKa = 11.84PRR53 pKa = 11.84SPHH56 pKa = 5.61RR57 pKa = 11.84PRR59 pKa = 11.84WVYY62 pKa = 10.86QLTSVWMVFVVVASILTNGLVLAATMKK89 pKa = 10.23FKK91 pKa = 11.0KK92 pKa = 9.7LRR94 pKa = 11.84HH95 pKa = 5.28PLNWILVNLAIADD108 pKa = 3.97LAEE111 pKa = 4.34TIIAGTISIVNQMYY125 pKa = 9.65GYY127 pKa = 10.28FVLGHH132 pKa = 6.48PLCIVEE138 pKa = 5.28GYY140 pKa = 7.23TVCLWGVIEE149 pKa = 4.02AHH151 pKa = 6.37LL152 pKa = 4.08
Molecular weight: 17.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3464131 |
50 |
4033 |
319.1 |
35.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.451 ± 0.026 | 2.26 ± 0.016 |
4.705 ± 0.017 | 6.972 ± 0.032 |
3.642 ± 0.018 | 6.82 ± 0.027 |
2.632 ± 0.013 | 3.93 ± 0.022 |
5.24 ± 0.033 | 10.364 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.1 ± 0.011 | 3.225 ± 0.016 |
6.551 ± 0.038 | 4.72 ± 0.022 |
5.911 ± 0.023 | 8.245 ± 0.034 |
5.14 ± 0.018 | 6.249 ± 0.018 |
1.252 ± 0.009 | 2.593 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
