
Wild carrot mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus; unclassified Umbravirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
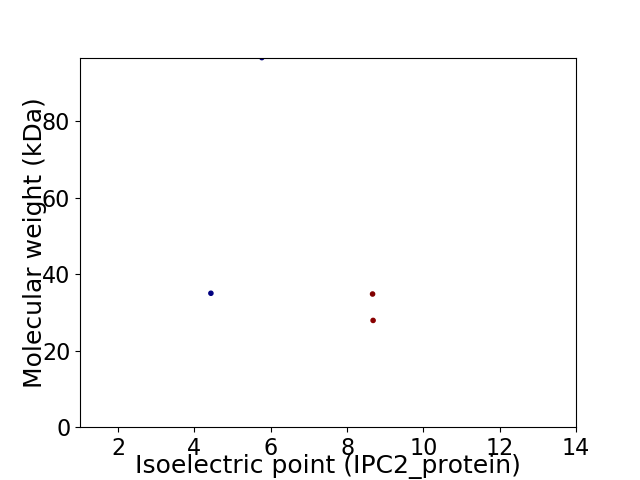
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

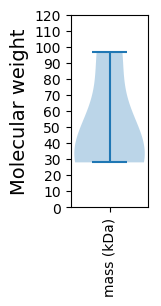
Protein with the lowest isoelectric point:
>tr|A0A6H2MW55|A0A6H2MW55_9TOMB RNA-directed RNA polymerase OS=Wild carrot mottle virus OX=1862686 PE=4 SV=1
MM1 pKa = 7.52SILRR5 pKa = 11.84KK6 pKa = 9.56VVQFLDD12 pKa = 3.79AKK14 pKa = 10.19YY15 pKa = 10.46SPAPGILSRR24 pKa = 11.84EE25 pKa = 4.09QCAEE29 pKa = 3.8RR30 pKa = 11.84YY31 pKa = 8.56GADD34 pKa = 2.8WAVLVTQEE42 pKa = 3.95RR43 pKa = 11.84MTRR46 pKa = 11.84ALSEE50 pKa = 4.04EE51 pKa = 3.93HH52 pKa = 6.45EE53 pKa = 4.24RR54 pKa = 11.84WYY56 pKa = 11.07SCNAEE61 pKa = 4.19TNVYY65 pKa = 10.09IPVTPYY71 pKa = 10.14VDD73 pKa = 3.48PRR75 pKa = 11.84DD76 pKa = 3.7VVAPLIEE83 pKa = 4.96DD84 pKa = 3.68AASTGDD90 pKa = 3.69AAPEE94 pKa = 3.96SDD96 pKa = 4.57AGEE99 pKa = 4.34PDD101 pKa = 3.17AAQLVGEE108 pKa = 4.65VVASRR113 pKa = 11.84PATGPGPTPYY123 pKa = 10.35SSEE126 pKa = 4.15EE127 pKa = 4.22LEE129 pKa = 4.51SATPYY134 pKa = 10.43CFTVEE139 pKa = 4.11AVGHH143 pKa = 6.67IIPVGRR149 pKa = 11.84MEE151 pKa = 4.34GTSVTRR157 pKa = 11.84EE158 pKa = 3.6GDD160 pKa = 3.4TAEE163 pKa = 4.67SDD165 pKa = 4.21SEE167 pKa = 4.36TDD169 pKa = 3.04TCSPAFYY176 pKa = 9.91IGAGNAVPVTAANGEE191 pKa = 4.14EE192 pKa = 4.53GQPQTATPIPGEE204 pKa = 4.22VPAFRR209 pKa = 11.84AEE211 pKa = 4.23DD212 pKa = 3.71PTISRR217 pKa = 11.84VPTTIVTGDD226 pKa = 3.42AAEE229 pKa = 4.25AMPTKK234 pKa = 10.57VDD236 pKa = 3.64DD237 pKa = 4.29CDD239 pKa = 4.75GSVSSSLMKK248 pKa = 10.63AGPTMAAMDD257 pKa = 4.29SYY259 pKa = 11.09SVAMEE264 pKa = 3.74LRR266 pKa = 11.84ARR268 pKa = 11.84FGLRR272 pKa = 11.84SKK274 pKa = 9.8TPANAEE280 pKa = 3.64LGGRR284 pKa = 11.84VSRR287 pKa = 11.84EE288 pKa = 3.8ILSHH292 pKa = 6.09CGGVRR297 pKa = 11.84SEE299 pKa = 4.29VYY301 pKa = 10.62YY302 pKa = 10.65LAQLAVQLWFAPTLLDD318 pKa = 4.9LVLTHH323 pKa = 6.04QVKK326 pKa = 10.6DD327 pKa = 3.86FCC329 pKa = 6.6
MM1 pKa = 7.52SILRR5 pKa = 11.84KK6 pKa = 9.56VVQFLDD12 pKa = 3.79AKK14 pKa = 10.19YY15 pKa = 10.46SPAPGILSRR24 pKa = 11.84EE25 pKa = 4.09QCAEE29 pKa = 3.8RR30 pKa = 11.84YY31 pKa = 8.56GADD34 pKa = 2.8WAVLVTQEE42 pKa = 3.95RR43 pKa = 11.84MTRR46 pKa = 11.84ALSEE50 pKa = 4.04EE51 pKa = 3.93HH52 pKa = 6.45EE53 pKa = 4.24RR54 pKa = 11.84WYY56 pKa = 11.07SCNAEE61 pKa = 4.19TNVYY65 pKa = 10.09IPVTPYY71 pKa = 10.14VDD73 pKa = 3.48PRR75 pKa = 11.84DD76 pKa = 3.7VVAPLIEE83 pKa = 4.96DD84 pKa = 3.68AASTGDD90 pKa = 3.69AAPEE94 pKa = 3.96SDD96 pKa = 4.57AGEE99 pKa = 4.34PDD101 pKa = 3.17AAQLVGEE108 pKa = 4.65VVASRR113 pKa = 11.84PATGPGPTPYY123 pKa = 10.35SSEE126 pKa = 4.15EE127 pKa = 4.22LEE129 pKa = 4.51SATPYY134 pKa = 10.43CFTVEE139 pKa = 4.11AVGHH143 pKa = 6.67IIPVGRR149 pKa = 11.84MEE151 pKa = 4.34GTSVTRR157 pKa = 11.84EE158 pKa = 3.6GDD160 pKa = 3.4TAEE163 pKa = 4.67SDD165 pKa = 4.21SEE167 pKa = 4.36TDD169 pKa = 3.04TCSPAFYY176 pKa = 9.91IGAGNAVPVTAANGEE191 pKa = 4.14EE192 pKa = 4.53GQPQTATPIPGEE204 pKa = 4.22VPAFRR209 pKa = 11.84AEE211 pKa = 4.23DD212 pKa = 3.71PTISRR217 pKa = 11.84VPTTIVTGDD226 pKa = 3.42AAEE229 pKa = 4.25AMPTKK234 pKa = 10.57VDD236 pKa = 3.64DD237 pKa = 4.29CDD239 pKa = 4.75GSVSSSLMKK248 pKa = 10.63AGPTMAAMDD257 pKa = 4.29SYY259 pKa = 11.09SVAMEE264 pKa = 3.74LRR266 pKa = 11.84ARR268 pKa = 11.84FGLRR272 pKa = 11.84SKK274 pKa = 9.8TPANAEE280 pKa = 3.64LGGRR284 pKa = 11.84VSRR287 pKa = 11.84EE288 pKa = 3.8ILSHH292 pKa = 6.09CGGVRR297 pKa = 11.84SEE299 pKa = 4.29VYY301 pKa = 10.62YY302 pKa = 10.65LAQLAVQLWFAPTLLDD318 pKa = 4.9LVLTHH323 pKa = 6.04QVKK326 pKa = 10.6DD327 pKa = 3.86FCC329 pKa = 6.6
Molecular weight: 35.01 kDa
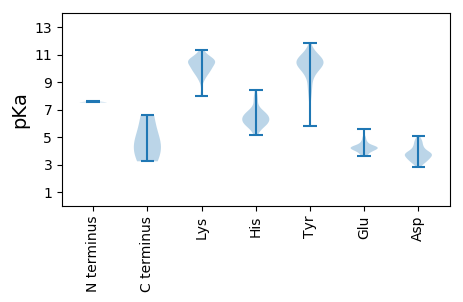
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H2MW43|A0A6H2MW43_9TOMB Cell to Cell MP OS=Wild carrot mottle virus OX=1862686 PE=4 SV=1
MM1 pKa = 7.63SGPGTPGFCPLWARR15 pKa = 11.84QVAHH19 pKa = 6.39TLSCKK24 pKa = 10.45LGALVIGIAALVSSGGACPQYY45 pKa = 10.81SHH47 pKa = 7.72PSGSRR52 pKa = 11.84QCRR55 pKa = 11.84EE56 pKa = 4.05GNSMTFRR63 pKa = 11.84LIYY66 pKa = 10.43DD67 pKa = 4.65GIEE70 pKa = 3.76QNSINNRR77 pKa = 11.84RR78 pKa = 11.84TNQPVTWRR86 pKa = 11.84SYY88 pKa = 10.33SGRR91 pKa = 11.84AQGARR96 pKa = 11.84PRR98 pKa = 11.84SYY100 pKa = 9.75DD101 pKa = 3.26TSPRR105 pKa = 11.84EE106 pKa = 3.97QYY108 pKa = 10.69GYY110 pKa = 9.31PHH112 pKa = 7.7AIIPAPPPAHH122 pKa = 5.87YY123 pKa = 10.29QPIPYY128 pKa = 8.75WPLAYY133 pKa = 7.61EE134 pKa = 4.17THH136 pKa = 6.63GGNAVYY142 pKa = 10.34RR143 pKa = 11.84EE144 pKa = 4.36VGGSVYY150 pKa = 10.47SPRR153 pKa = 11.84ARR155 pKa = 11.84HH156 pKa = 5.14GRR158 pKa = 11.84GRR160 pKa = 11.84GNYY163 pKa = 8.08MGARR167 pKa = 11.84HH168 pKa = 5.77SSSRR172 pKa = 11.84TGANRR177 pKa = 11.84GEE179 pKa = 4.29PEE181 pKa = 4.08SLHH184 pKa = 5.59TFVFRR189 pKa = 11.84TPAGRR194 pKa = 11.84FLSEE198 pKa = 4.34LFNSVEE204 pKa = 4.07RR205 pKa = 11.84LSRR208 pKa = 11.84EE209 pKa = 4.24CPSLLQSSDD218 pKa = 3.42PVGGGSTGARR228 pKa = 11.84GEE230 pKa = 4.11RR231 pKa = 11.84LLAVLNVAANNRR243 pKa = 11.84GEE245 pKa = 4.32GAVLQANTTGGCGNPEE261 pKa = 4.0AQHH264 pKa = 5.84QVTTPRR270 pKa = 11.84PNPTAAIPEE279 pKa = 4.12CRR281 pKa = 11.84IDD283 pKa = 3.3EE284 pKa = 4.36RR285 pKa = 11.84GRR287 pKa = 11.84DD288 pKa = 3.78GEE290 pKa = 4.43GTHH293 pKa = 6.42TKK295 pKa = 10.36LAVPHH300 pKa = 6.12PHH302 pKa = 6.18LCGRR306 pKa = 11.84VHH308 pKa = 7.96SEE310 pKa = 4.03GEE312 pKa = 4.08PCQGEE317 pKa = 4.1PTVRR321 pKa = 11.84YY322 pKa = 9.25LL323 pKa = 4.3
MM1 pKa = 7.63SGPGTPGFCPLWARR15 pKa = 11.84QVAHH19 pKa = 6.39TLSCKK24 pKa = 10.45LGALVIGIAALVSSGGACPQYY45 pKa = 10.81SHH47 pKa = 7.72PSGSRR52 pKa = 11.84QCRR55 pKa = 11.84EE56 pKa = 4.05GNSMTFRR63 pKa = 11.84LIYY66 pKa = 10.43DD67 pKa = 4.65GIEE70 pKa = 3.76QNSINNRR77 pKa = 11.84RR78 pKa = 11.84TNQPVTWRR86 pKa = 11.84SYY88 pKa = 10.33SGRR91 pKa = 11.84AQGARR96 pKa = 11.84PRR98 pKa = 11.84SYY100 pKa = 9.75DD101 pKa = 3.26TSPRR105 pKa = 11.84EE106 pKa = 3.97QYY108 pKa = 10.69GYY110 pKa = 9.31PHH112 pKa = 7.7AIIPAPPPAHH122 pKa = 5.87YY123 pKa = 10.29QPIPYY128 pKa = 8.75WPLAYY133 pKa = 7.61EE134 pKa = 4.17THH136 pKa = 6.63GGNAVYY142 pKa = 10.34RR143 pKa = 11.84EE144 pKa = 4.36VGGSVYY150 pKa = 10.47SPRR153 pKa = 11.84ARR155 pKa = 11.84HH156 pKa = 5.14GRR158 pKa = 11.84GRR160 pKa = 11.84GNYY163 pKa = 8.08MGARR167 pKa = 11.84HH168 pKa = 5.77SSSRR172 pKa = 11.84TGANRR177 pKa = 11.84GEE179 pKa = 4.29PEE181 pKa = 4.08SLHH184 pKa = 5.59TFVFRR189 pKa = 11.84TPAGRR194 pKa = 11.84FLSEE198 pKa = 4.34LFNSVEE204 pKa = 4.07RR205 pKa = 11.84LSRR208 pKa = 11.84EE209 pKa = 4.24CPSLLQSSDD218 pKa = 3.42PVGGGSTGARR228 pKa = 11.84GEE230 pKa = 4.11RR231 pKa = 11.84LLAVLNVAANNRR243 pKa = 11.84GEE245 pKa = 4.32GAVLQANTTGGCGNPEE261 pKa = 4.0AQHH264 pKa = 5.84QVTTPRR270 pKa = 11.84PNPTAAIPEE279 pKa = 4.12CRR281 pKa = 11.84IDD283 pKa = 3.3EE284 pKa = 4.36RR285 pKa = 11.84GRR287 pKa = 11.84DD288 pKa = 3.78GEE290 pKa = 4.43GTHH293 pKa = 6.42TKK295 pKa = 10.36LAVPHH300 pKa = 6.12PHH302 pKa = 6.18LCGRR306 pKa = 11.84VHH308 pKa = 7.96SEE310 pKa = 4.03GEE312 pKa = 4.08PCQGEE317 pKa = 4.1PTVRR321 pKa = 11.84YY322 pKa = 9.25LL323 pKa = 4.3
Molecular weight: 34.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1779 |
253 |
874 |
444.8 |
48.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.162 ± 0.8 | 2.024 ± 0.255 |
4.609 ± 1.008 | 6.352 ± 0.542 |
2.923 ± 0.609 | 8.094 ± 1.033 |
2.361 ± 0.376 | 4.16 ± 0.45 |
3.316 ± 1.077 | 7.532 ± 0.716 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.248 ± 0.43 | 2.923 ± 0.416 |
7.701 ± 0.559 | 3.373 ± 0.274 |
6.633 ± 0.862 | 7.195 ± 0.513 |
6.633 ± 0.851 | 7.87 ± 0.667 |
1.124 ± 0.095 | 3.766 ± 0.118 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
