
Sparassis crispa
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Sparassidaceae; Sparassis
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
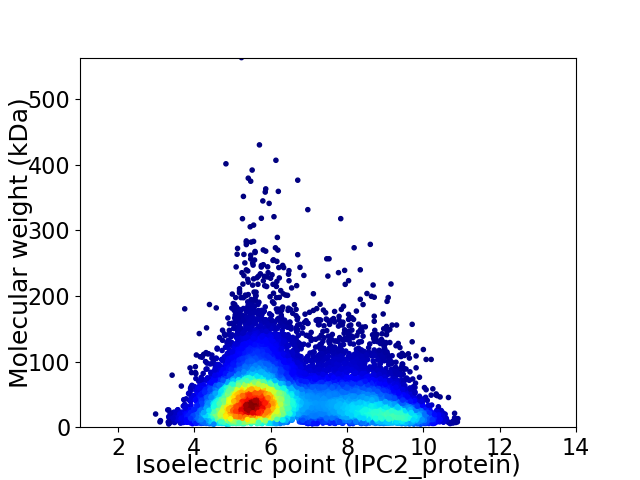
Virtual 2D-PAGE plot for 13078 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A401GL46|A0A401GL46_9APHY Non-specific serine/threonine protein kinase OS=Sparassis crispa OX=139825 GN=SCP_0412730 PE=4 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84SKK5 pKa = 10.7LQSAVGGGAKK15 pKa = 10.35DD16 pKa = 3.25STSLHH21 pKa = 5.62RR22 pKa = 11.84WVLLKK27 pKa = 10.99NSIVRR32 pKa = 11.84SSPSDD37 pKa = 3.08ATTATPEE44 pKa = 4.33GADD47 pKa = 3.4VTDD50 pKa = 4.62VYY52 pKa = 11.15PHH54 pKa = 7.4DD55 pKa = 4.71DD56 pKa = 3.99EE57 pKa = 5.57EE58 pKa = 4.52VHH60 pKa = 6.8EE61 pKa = 5.83DD62 pKa = 3.61EE63 pKa = 6.62DD64 pKa = 4.22SDD66 pKa = 5.41AFMFPDD72 pKa = 4.11PDD74 pKa = 3.46AVRR77 pKa = 11.84GSRR80 pKa = 11.84DD81 pKa = 3.09SGLCDD86 pKa = 5.46PEE88 pKa = 4.36NQWLDD93 pKa = 3.55SLLQTLGDD101 pKa = 4.1DD102 pKa = 4.12DD103 pKa = 3.94QAEE106 pKa = 4.41SEE108 pKa = 4.79VPVNAADD115 pKa = 5.7DD116 pKa = 4.4DD117 pKa = 4.59DD118 pKa = 5.5EE119 pKa = 4.92PLSPLCSPMSSSDD132 pKa = 3.39DD133 pKa = 3.72LVNHH137 pKa = 6.33SSYY140 pKa = 11.77YY141 pKa = 10.25EE142 pKa = 3.84IQPPLSIPYY151 pKa = 8.16PVPYY155 pKa = 9.92PPLHH159 pKa = 6.73PPLVPAWFEE168 pKa = 4.28DD169 pKa = 4.06EE170 pKa = 4.66SPSDD174 pKa = 3.77SLLDD178 pKa = 3.69SSPPLYY184 pKa = 10.35HH185 pKa = 7.72DD186 pKa = 4.25PLPYY190 pKa = 10.42YY191 pKa = 10.67DD192 pKa = 4.92VDD194 pKa = 3.82DD195 pKa = 4.72TEE197 pKa = 5.67DD198 pKa = 3.81LAVPDD203 pKa = 5.32AIEE206 pKa = 4.57DD207 pKa = 3.95TSDD210 pKa = 4.1DD211 pKa = 3.89EE212 pKa = 5.66SDD214 pKa = 4.03APSTPCYY221 pKa = 10.23RR222 pKa = 11.84STTSLSPTDD231 pKa = 3.79PASIPLPPEE240 pKa = 3.53QTRR243 pKa = 11.84LRR245 pKa = 11.84SHH247 pKa = 7.4PLQPRR252 pKa = 11.84VYY254 pKa = 9.95IDD256 pKa = 3.33TDD258 pKa = 3.16DD259 pKa = 3.97SYY261 pKa = 11.71FYY263 pKa = 10.65PFDD266 pKa = 3.82LTSMPSHH273 pKa = 7.6DD274 pKa = 4.96DD275 pKa = 3.27DD276 pKa = 5.58HH277 pKa = 7.12PISARR282 pKa = 11.84VFQPPIYY289 pKa = 9.9QEE291 pKa = 4.2CC292 pKa = 3.48
MM1 pKa = 7.51ARR3 pKa = 11.84SKK5 pKa = 10.7LQSAVGGGAKK15 pKa = 10.35DD16 pKa = 3.25STSLHH21 pKa = 5.62RR22 pKa = 11.84WVLLKK27 pKa = 10.99NSIVRR32 pKa = 11.84SSPSDD37 pKa = 3.08ATTATPEE44 pKa = 4.33GADD47 pKa = 3.4VTDD50 pKa = 4.62VYY52 pKa = 11.15PHH54 pKa = 7.4DD55 pKa = 4.71DD56 pKa = 3.99EE57 pKa = 5.57EE58 pKa = 4.52VHH60 pKa = 6.8EE61 pKa = 5.83DD62 pKa = 3.61EE63 pKa = 6.62DD64 pKa = 4.22SDD66 pKa = 5.41AFMFPDD72 pKa = 4.11PDD74 pKa = 3.46AVRR77 pKa = 11.84GSRR80 pKa = 11.84DD81 pKa = 3.09SGLCDD86 pKa = 5.46PEE88 pKa = 4.36NQWLDD93 pKa = 3.55SLLQTLGDD101 pKa = 4.1DD102 pKa = 4.12DD103 pKa = 3.94QAEE106 pKa = 4.41SEE108 pKa = 4.79VPVNAADD115 pKa = 5.7DD116 pKa = 4.4DD117 pKa = 4.59DD118 pKa = 5.5EE119 pKa = 4.92PLSPLCSPMSSSDD132 pKa = 3.39DD133 pKa = 3.72LVNHH137 pKa = 6.33SSYY140 pKa = 11.77YY141 pKa = 10.25EE142 pKa = 3.84IQPPLSIPYY151 pKa = 8.16PVPYY155 pKa = 9.92PPLHH159 pKa = 6.73PPLVPAWFEE168 pKa = 4.28DD169 pKa = 4.06EE170 pKa = 4.66SPSDD174 pKa = 3.77SLLDD178 pKa = 3.69SSPPLYY184 pKa = 10.35HH185 pKa = 7.72DD186 pKa = 4.25PLPYY190 pKa = 10.42YY191 pKa = 10.67DD192 pKa = 4.92VDD194 pKa = 3.82DD195 pKa = 4.72TEE197 pKa = 5.67DD198 pKa = 3.81LAVPDD203 pKa = 5.32AIEE206 pKa = 4.57DD207 pKa = 3.95TSDD210 pKa = 4.1DD211 pKa = 3.89EE212 pKa = 5.66SDD214 pKa = 4.03APSTPCYY221 pKa = 10.23RR222 pKa = 11.84STTSLSPTDD231 pKa = 3.79PASIPLPPEE240 pKa = 3.53QTRR243 pKa = 11.84LRR245 pKa = 11.84SHH247 pKa = 7.4PLQPRR252 pKa = 11.84VYY254 pKa = 9.95IDD256 pKa = 3.33TDD258 pKa = 3.16DD259 pKa = 3.97SYY261 pKa = 11.71FYY263 pKa = 10.65PFDD266 pKa = 3.82LTSMPSHH273 pKa = 7.6DD274 pKa = 4.96DD275 pKa = 3.27DD276 pKa = 5.58HH277 pKa = 7.12PISARR282 pKa = 11.84VFQPPIYY289 pKa = 9.9QEE291 pKa = 4.2CC292 pKa = 3.48
Molecular weight: 32.21 kDa
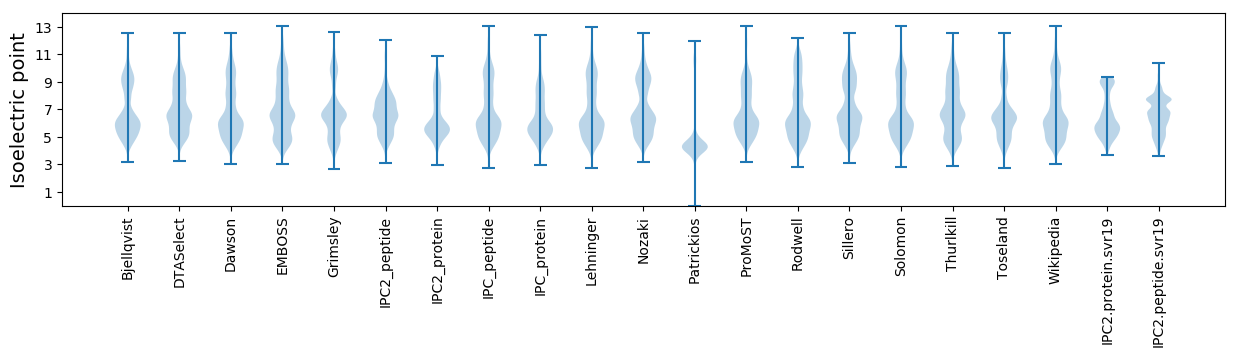
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A401GVM3|A0A401GVM3_9APHY Glycerol kinase OS=Sparassis crispa OX=139825 GN=SCP_0901190 PE=3 SV=1
MM1 pKa = 7.98RR2 pKa = 11.84KK3 pKa = 9.0PLQQRR8 pKa = 11.84VGTLGRR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84KK18 pKa = 9.04SAFRR22 pKa = 11.84LPSNSAVPIPSNPAISAQARR42 pKa = 11.84VLASLTRR49 pKa = 11.84LASFSAPAFIAAGALSAGTQSAPRR73 pKa = 11.84GFPDD77 pKa = 3.68PVKK80 pKa = 10.14PHH82 pKa = 6.06HH83 pKa = 6.31TPPRR87 pKa = 11.84LPAPFLDD94 pKa = 4.58LPRR97 pKa = 11.84HH98 pKa = 4.66VQAHH102 pKa = 5.88RR103 pKa = 11.84SQRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84SRR112 pKa = 11.84TGHH115 pKa = 6.74LGATTACSSLLAVTQAPTKK134 pKa = 10.44IPTLPSTLVAFAHH147 pKa = 6.41CGEE150 pKa = 4.54RR151 pKa = 11.84PHH153 pKa = 6.73PGRR156 pKa = 11.84RR157 pKa = 11.84AGHH160 pKa = 6.57RR161 pKa = 11.84GATTAPSPSSRR172 pKa = 11.84PPSPRR177 pKa = 11.84RR178 pKa = 11.84GPSSPLRR185 pKa = 11.84TLALPSSTRR194 pKa = 11.84VRR196 pKa = 11.84PLRR199 pKa = 3.94
MM1 pKa = 7.98RR2 pKa = 11.84KK3 pKa = 9.0PLQQRR8 pKa = 11.84VGTLGRR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84KK18 pKa = 9.04SAFRR22 pKa = 11.84LPSNSAVPIPSNPAISAQARR42 pKa = 11.84VLASLTRR49 pKa = 11.84LASFSAPAFIAAGALSAGTQSAPRR73 pKa = 11.84GFPDD77 pKa = 3.68PVKK80 pKa = 10.14PHH82 pKa = 6.06HH83 pKa = 6.31TPPRR87 pKa = 11.84LPAPFLDD94 pKa = 4.58LPRR97 pKa = 11.84HH98 pKa = 4.66VQAHH102 pKa = 5.88RR103 pKa = 11.84SQRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84SRR112 pKa = 11.84TGHH115 pKa = 6.74LGATTACSSLLAVTQAPTKK134 pKa = 10.44IPTLPSTLVAFAHH147 pKa = 6.41CGEE150 pKa = 4.54RR151 pKa = 11.84PHH153 pKa = 6.73PGRR156 pKa = 11.84RR157 pKa = 11.84AGHH160 pKa = 6.57RR161 pKa = 11.84GATTAPSPSSRR172 pKa = 11.84PPSPRR177 pKa = 11.84RR178 pKa = 11.84GPSSPLRR185 pKa = 11.84TLALPSSTRR194 pKa = 11.84VRR196 pKa = 11.84PLRR199 pKa = 3.94
Molecular weight: 21.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5783113 |
49 |
5044 |
442.2 |
48.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.85 ± 0.019 | 1.301 ± 0.01 |
5.751 ± 0.016 | 6.085 ± 0.022 |
3.565 ± 0.014 | 6.261 ± 0.018 |
2.71 ± 0.011 | 4.553 ± 0.015 |
4.364 ± 0.02 | 9.17 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.008 | 3.188 ± 0.011 |
6.707 ± 0.025 | 3.704 ± 0.013 |
6.485 ± 0.016 | 8.604 ± 0.033 |
5.871 ± 0.013 | 6.564 ± 0.016 |
1.421 ± 0.008 | 2.631 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
