
Rhizobium subbaraonis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
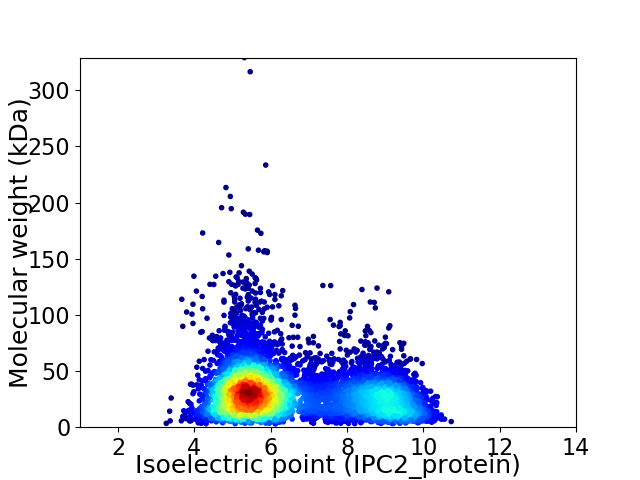
Virtual 2D-PAGE plot for 6271 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285UYQ2|A0A285UYQ2_9RHIZ Type IV secretion system protein VirB3 OS=Rhizobium subbaraonis OX=908946 GN=SAMN05892877_12543 PE=4 SV=1
MM1 pKa = 7.06GVRR4 pKa = 11.84YY5 pKa = 9.88VKK7 pKa = 10.71YY8 pKa = 8.84RR9 pKa = 11.84TDD11 pKa = 3.12TGEE14 pKa = 4.07IIATGTCSNQEE25 pKa = 3.74HH26 pKa = 7.1CFLQMEE32 pKa = 4.5DD33 pKa = 4.32AEE35 pKa = 4.56NEE37 pKa = 4.29HH38 pKa = 6.8ILLDD42 pKa = 3.89VSADD46 pKa = 5.0DD47 pKa = 3.55ATQWIDD53 pKa = 3.92DD54 pKa = 3.81QGEE57 pKa = 4.01LADD60 pKa = 5.57RR61 pKa = 11.84PVLDD65 pKa = 5.02LPTATSIAADD75 pKa = 4.98GIAEE79 pKa = 4.32VVGALPAGTVVRR91 pKa = 11.84FNGAAATTDD100 pKa = 3.22GAAFAFTTDD109 pKa = 3.04MPGTYY114 pKa = 10.12VFDD117 pKa = 4.48FEE119 pKa = 5.38PPFPYY124 pKa = 9.92IPCTLTVEE132 pKa = 4.25ATDD135 pKa = 4.98AII137 pKa = 4.34
MM1 pKa = 7.06GVRR4 pKa = 11.84YY5 pKa = 9.88VKK7 pKa = 10.71YY8 pKa = 8.84RR9 pKa = 11.84TDD11 pKa = 3.12TGEE14 pKa = 4.07IIATGTCSNQEE25 pKa = 3.74HH26 pKa = 7.1CFLQMEE32 pKa = 4.5DD33 pKa = 4.32AEE35 pKa = 4.56NEE37 pKa = 4.29HH38 pKa = 6.8ILLDD42 pKa = 3.89VSADD46 pKa = 5.0DD47 pKa = 3.55ATQWIDD53 pKa = 3.92DD54 pKa = 3.81QGEE57 pKa = 4.01LADD60 pKa = 5.57RR61 pKa = 11.84PVLDD65 pKa = 5.02LPTATSIAADD75 pKa = 4.98GIAEE79 pKa = 4.32VVGALPAGTVVRR91 pKa = 11.84FNGAAATTDD100 pKa = 3.22GAAFAFTTDD109 pKa = 3.04MPGTYY114 pKa = 10.12VFDD117 pKa = 4.48FEE119 pKa = 5.38PPFPYY124 pKa = 9.92IPCTLTVEE132 pKa = 4.25ATDD135 pKa = 4.98AII137 pKa = 4.34
Molecular weight: 14.62 kDa
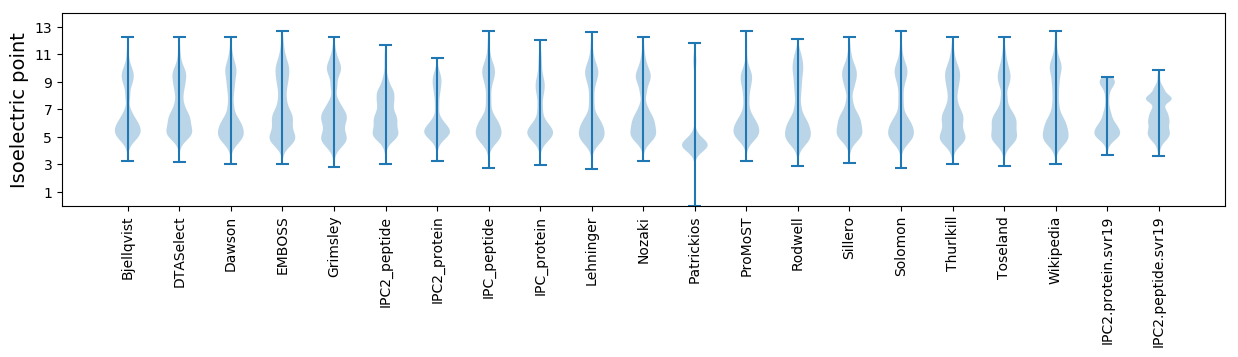
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285V268|A0A285V268_9RHIZ Histidine ammonia-lyase OS=Rhizobium subbaraonis OX=908946 GN=SAMN05892877_14414 PE=4 SV=1
MM1 pKa = 7.63SPEE4 pKa = 3.96RR5 pKa = 11.84SKK7 pKa = 11.39LPWIWVVKK15 pKa = 8.73PPRR18 pKa = 11.84VVPPPGLFAPFCSGRR33 pKa = 11.84RR34 pKa = 11.84DD35 pKa = 4.28SRR37 pKa = 11.84EE38 pKa = 3.36QWSNRR43 pKa = 11.84TSEE46 pKa = 4.04SGAPMGAKK54 pKa = 9.48VSKK57 pKa = 10.04KK58 pKa = 9.74ASTRR62 pKa = 11.84RR63 pKa = 11.84PCSADD68 pKa = 3.14RR69 pKa = 11.84SAPRR73 pKa = 11.84PSSICSDD80 pKa = 3.14RR81 pKa = 11.84RR82 pKa = 11.84DD83 pKa = 3.47AGGLGISIRR92 pKa = 11.84WACKK96 pKa = 8.06VLRR99 pKa = 11.84FDD101 pKa = 3.68TSTYY105 pKa = 9.27HH106 pKa = 5.16YY107 pKa = 10.04KK108 pKa = 10.26SRR110 pKa = 11.84RR111 pKa = 11.84TGQAPLEE118 pKa = 4.09RR119 pKa = 11.84RR120 pKa = 11.84IKK122 pKa = 10.13EE123 pKa = 4.27YY124 pKa = 10.76VRR126 pKa = 11.84HH127 pKa = 5.59ACGMATAVRR136 pKa = 11.84MVTCAAMDD144 pKa = 3.66GRR146 pKa = 11.84STINMKK152 pKa = 8.54MRR154 pKa = 11.84RR155 pKa = 11.84FRR157 pKa = 11.84LFIDD161 pKa = 3.89RR162 pKa = 11.84CCLYY166 pKa = 10.95VADD169 pKa = 4.85SPKK172 pKa = 10.55KK173 pKa = 10.47LQILLQINSSEE184 pKa = 4.09CRR186 pKa = 11.84SADD189 pKa = 3.26VAFVHH194 pKa = 6.99FKK196 pKa = 10.87RR197 pKa = 11.84SIHH200 pKa = 5.7TAMQTLSGEE209 pKa = 4.28SSGLYY214 pKa = 9.58QCFLRR219 pKa = 11.84ADD221 pKa = 3.46TDD223 pKa = 4.11YY224 pKa = 11.26ILPRR228 pKa = 11.84DD229 pKa = 3.42IYY231 pKa = 10.91QRR233 pKa = 11.84TRR235 pKa = 11.84SPLL238 pKa = 3.33
MM1 pKa = 7.63SPEE4 pKa = 3.96RR5 pKa = 11.84SKK7 pKa = 11.39LPWIWVVKK15 pKa = 8.73PPRR18 pKa = 11.84VVPPPGLFAPFCSGRR33 pKa = 11.84RR34 pKa = 11.84DD35 pKa = 4.28SRR37 pKa = 11.84EE38 pKa = 3.36QWSNRR43 pKa = 11.84TSEE46 pKa = 4.04SGAPMGAKK54 pKa = 9.48VSKK57 pKa = 10.04KK58 pKa = 9.74ASTRR62 pKa = 11.84RR63 pKa = 11.84PCSADD68 pKa = 3.14RR69 pKa = 11.84SAPRR73 pKa = 11.84PSSICSDD80 pKa = 3.14RR81 pKa = 11.84RR82 pKa = 11.84DD83 pKa = 3.47AGGLGISIRR92 pKa = 11.84WACKK96 pKa = 8.06VLRR99 pKa = 11.84FDD101 pKa = 3.68TSTYY105 pKa = 9.27HH106 pKa = 5.16YY107 pKa = 10.04KK108 pKa = 10.26SRR110 pKa = 11.84RR111 pKa = 11.84TGQAPLEE118 pKa = 4.09RR119 pKa = 11.84RR120 pKa = 11.84IKK122 pKa = 10.13EE123 pKa = 4.27YY124 pKa = 10.76VRR126 pKa = 11.84HH127 pKa = 5.59ACGMATAVRR136 pKa = 11.84MVTCAAMDD144 pKa = 3.66GRR146 pKa = 11.84STINMKK152 pKa = 8.54MRR154 pKa = 11.84RR155 pKa = 11.84FRR157 pKa = 11.84LFIDD161 pKa = 3.89RR162 pKa = 11.84CCLYY166 pKa = 10.95VADD169 pKa = 4.85SPKK172 pKa = 10.55KK173 pKa = 10.47LQILLQINSSEE184 pKa = 4.09CRR186 pKa = 11.84SADD189 pKa = 3.26VAFVHH194 pKa = 6.99FKK196 pKa = 10.87RR197 pKa = 11.84SIHH200 pKa = 5.7TAMQTLSGEE209 pKa = 4.28SSGLYY214 pKa = 9.58QCFLRR219 pKa = 11.84ADD221 pKa = 3.46TDD223 pKa = 4.11YY224 pKa = 11.26ILPRR228 pKa = 11.84DD229 pKa = 3.42IYY231 pKa = 10.91QRR233 pKa = 11.84TRR235 pKa = 11.84SPLL238 pKa = 3.33
Molecular weight: 27.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1918300 |
27 |
3078 |
305.9 |
33.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.268 ± 0.039 | 0.814 ± 0.01 |
5.743 ± 0.026 | 5.867 ± 0.03 |
3.83 ± 0.023 | 8.497 ± 0.03 |
2.007 ± 0.016 | 5.416 ± 0.024 |
3.447 ± 0.025 | 9.881 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.014 | 2.7 ± 0.016 |
4.905 ± 0.025 | 3.071 ± 0.017 |
7.101 ± 0.037 | 5.637 ± 0.021 |
5.32 ± 0.022 | 7.389 ± 0.027 |
1.302 ± 0.012 | 2.272 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
