
Haloarcula marismortui (strain ATCC 43049 / DSM 3752 / JCM 8966 / VKM B-1809) (Halobacterium marismortui)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Haloarcula; Haloarcula marismortui
Average proteome isoelectric point is 4.91
Get precalculated fractions of proteins
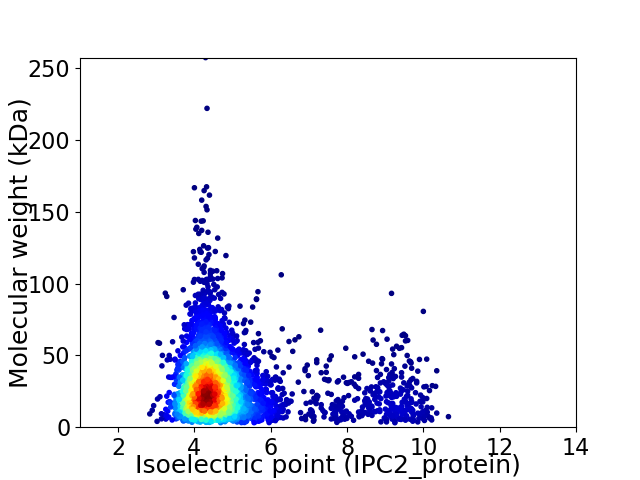
Virtual 2D-PAGE plot for 4234 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
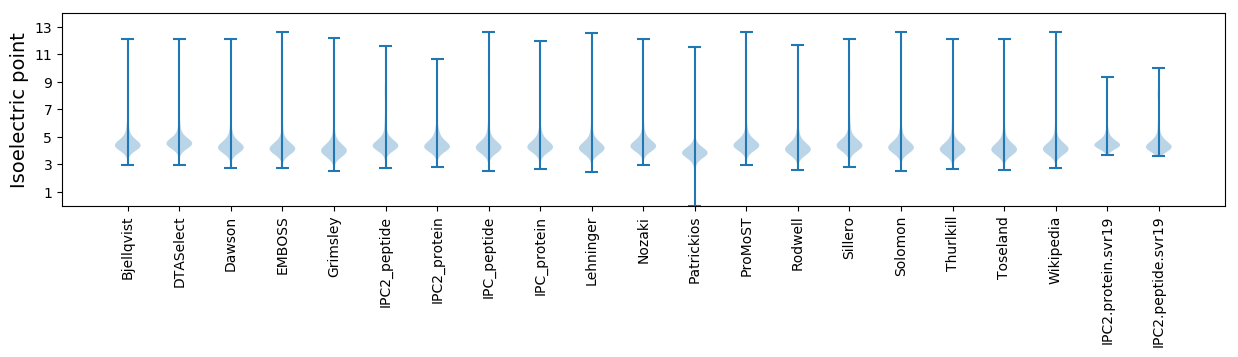
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5UWZ5|Q5UWZ5_HALMA Uncharacterized protein OS=Haloarcula marismortui (strain ATCC 43049 / DSM 3752 / JCM 8966 / VKM B-1809) OX=272569 GN=rrnAC3542 PE=4 SV=1
MM1 pKa = 7.57SEE3 pKa = 3.97YY4 pKa = 10.66TGDD7 pKa = 3.25VTLNGGIEE15 pKa = 4.34TPVRR19 pKa = 11.84MSGLEE24 pKa = 3.92DD25 pKa = 3.91MYY27 pKa = 11.26VQPDD31 pKa = 4.38SIDD34 pKa = 3.32GDD36 pKa = 3.77LDD38 pKa = 3.74LRR40 pKa = 11.84NVEE43 pKa = 4.52YY44 pKa = 10.9VFTGVPITPAADD56 pKa = 3.22VADD59 pKa = 4.38PVTDD63 pKa = 3.21ITGTLEE69 pKa = 4.65DD70 pKa = 5.67GYY72 pKa = 9.36TEE74 pKa = 4.42PDD76 pKa = 3.77GVHH79 pKa = 6.79GDD81 pKa = 3.84LAVGTAEE88 pKa = 5.07DD89 pKa = 3.69VFIAHH94 pKa = 6.96GAVDD98 pKa = 4.05GALSARR104 pKa = 11.84GPEE107 pKa = 3.96QVFDD111 pKa = 4.73AGTSDD116 pKa = 3.77APSRR120 pKa = 11.84DD121 pKa = 3.18PATYY125 pKa = 10.6DD126 pKa = 3.22VTVSGWQQQRR136 pKa = 11.84EE137 pKa = 4.35VTDD140 pKa = 4.06PNTGVVLSGCQSTVNVTGATGSVSCYY166 pKa = 10.71LIGTNNEE173 pKa = 4.11LIVRR177 pKa = 11.84GDD179 pKa = 3.36AAVEE183 pKa = 3.64VHH185 pKa = 6.68IVGRR189 pKa = 11.84DD190 pKa = 3.23NRR192 pKa = 11.84VDD194 pKa = 3.89LGPYY198 pKa = 9.43IDD200 pKa = 4.53VDD202 pKa = 3.94TAVEE206 pKa = 4.18TGFDD210 pKa = 3.5NTIDD214 pKa = 3.9VQPFPADD221 pKa = 3.56DD222 pKa = 4.88LIEE225 pKa = 4.3TTEE228 pKa = 4.2DD229 pKa = 3.08EE230 pKa = 4.79AYY232 pKa = 10.62SAVTFGRR239 pKa = 11.84AKK241 pKa = 9.3VTYY244 pKa = 9.95QSVAEE249 pKa = 4.39DD250 pKa = 3.93EE251 pKa = 4.42DD252 pKa = 3.66WCRR255 pKa = 11.84GCGEE259 pKa = 4.09AADD262 pKa = 5.06AIIEE266 pKa = 4.19RR267 pKa = 11.84QQKK270 pKa = 9.1EE271 pKa = 3.87AFFILGTPIITYY283 pKa = 9.89EE284 pKa = 4.08EE285 pKa = 4.62GGGSYY290 pKa = 10.06EE291 pKa = 4.73CEE293 pKa = 4.22HH294 pKa = 6.98CAHH297 pKa = 6.94SAPDD301 pKa = 3.49TSLTPEE307 pKa = 3.71EE308 pKa = 4.06RR309 pKa = 11.84RR310 pKa = 11.84EE311 pKa = 3.99IFF313 pKa = 3.63
MM1 pKa = 7.57SEE3 pKa = 3.97YY4 pKa = 10.66TGDD7 pKa = 3.25VTLNGGIEE15 pKa = 4.34TPVRR19 pKa = 11.84MSGLEE24 pKa = 3.92DD25 pKa = 3.91MYY27 pKa = 11.26VQPDD31 pKa = 4.38SIDD34 pKa = 3.32GDD36 pKa = 3.77LDD38 pKa = 3.74LRR40 pKa = 11.84NVEE43 pKa = 4.52YY44 pKa = 10.9VFTGVPITPAADD56 pKa = 3.22VADD59 pKa = 4.38PVTDD63 pKa = 3.21ITGTLEE69 pKa = 4.65DD70 pKa = 5.67GYY72 pKa = 9.36TEE74 pKa = 4.42PDD76 pKa = 3.77GVHH79 pKa = 6.79GDD81 pKa = 3.84LAVGTAEE88 pKa = 5.07DD89 pKa = 3.69VFIAHH94 pKa = 6.96GAVDD98 pKa = 4.05GALSARR104 pKa = 11.84GPEE107 pKa = 3.96QVFDD111 pKa = 4.73AGTSDD116 pKa = 3.77APSRR120 pKa = 11.84DD121 pKa = 3.18PATYY125 pKa = 10.6DD126 pKa = 3.22VTVSGWQQQRR136 pKa = 11.84EE137 pKa = 4.35VTDD140 pKa = 4.06PNTGVVLSGCQSTVNVTGATGSVSCYY166 pKa = 10.71LIGTNNEE173 pKa = 4.11LIVRR177 pKa = 11.84GDD179 pKa = 3.36AAVEE183 pKa = 3.64VHH185 pKa = 6.68IVGRR189 pKa = 11.84DD190 pKa = 3.23NRR192 pKa = 11.84VDD194 pKa = 3.89LGPYY198 pKa = 9.43IDD200 pKa = 4.53VDD202 pKa = 3.94TAVEE206 pKa = 4.18TGFDD210 pKa = 3.5NTIDD214 pKa = 3.9VQPFPADD221 pKa = 3.56DD222 pKa = 4.88LIEE225 pKa = 4.3TTEE228 pKa = 4.2DD229 pKa = 3.08EE230 pKa = 4.79AYY232 pKa = 10.62SAVTFGRR239 pKa = 11.84AKK241 pKa = 9.3VTYY244 pKa = 9.95QSVAEE249 pKa = 4.39DD250 pKa = 3.93EE251 pKa = 4.42DD252 pKa = 3.66WCRR255 pKa = 11.84GCGEE259 pKa = 4.09AADD262 pKa = 5.06AIIEE266 pKa = 4.19RR267 pKa = 11.84QQKK270 pKa = 9.1EE271 pKa = 3.87AFFILGTPIITYY283 pKa = 9.89EE284 pKa = 4.08EE285 pKa = 4.62GGGSYY290 pKa = 10.06EE291 pKa = 4.73CEE293 pKa = 4.22HH294 pKa = 6.98CAHH297 pKa = 6.94SAPDD301 pKa = 3.49TSLTPEE307 pKa = 3.71EE308 pKa = 4.06RR309 pKa = 11.84RR310 pKa = 11.84EE311 pKa = 3.99IFF313 pKa = 3.63
Molecular weight: 33.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5V814|Q5V814_HALMA MCM / cell division control protein 21 OS=Haloarcula marismortui (strain ATCC 43049 / DSM 3752 / JCM 8966 / VKM B-1809) OX=272569 GN=mcm3 PE=3 SV=1
MM1 pKa = 7.23QNTHH5 pKa = 7.18AIEE8 pKa = 3.98QKK10 pKa = 10.44GRR12 pKa = 11.84GIAVALRR19 pKa = 11.84PSPRR23 pKa = 11.84DD24 pKa = 3.17AGAGSGRR31 pKa = 11.84ASAGASVQPVPAPGALPAGHH51 pKa = 6.89PRR53 pKa = 11.84SHH55 pKa = 6.26EE56 pKa = 3.85VRR58 pKa = 11.84HH59 pKa = 5.27RR60 pKa = 11.84ASRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84TRR67 pKa = 11.84SAPAASFRR75 pKa = 11.84LRR77 pKa = 11.84RR78 pKa = 11.84SGPSPGAPGRR88 pKa = 11.84HH89 pKa = 6.15AGLAGGSRR97 pKa = 11.84PYY99 pKa = 10.53SEE101 pKa = 5.45RR102 pKa = 11.84SYY104 pKa = 11.68SCIQPPRR111 pKa = 11.84EE112 pKa = 4.09IVDD115 pKa = 3.32EE116 pKa = 4.2RR117 pKa = 11.84APDD120 pKa = 3.21VWANGPAAFVLRR132 pKa = 11.84ALAPPCEE139 pKa = 4.24CFIGRR144 pKa = 11.84VCEE147 pKa = 4.02FDD149 pKa = 3.94LLGAVVASDD158 pKa = 4.14DD159 pKa = 3.67GHH161 pKa = 7.24GEE163 pKa = 4.0LVRR166 pKa = 11.84VPVVGRR172 pKa = 11.84AFAVDD177 pKa = 4.07AQEE180 pKa = 3.25QWAALRR186 pKa = 11.84EE187 pKa = 4.24QFTGNTFLRR196 pKa = 11.84MCVSRR201 pKa = 11.84ADD203 pKa = 3.71RR204 pKa = 11.84FTSRR208 pKa = 11.84RR209 pKa = 11.84SRR211 pKa = 11.84LTHH214 pKa = 5.8GSQWNRR220 pKa = 11.84RR221 pKa = 11.84AWCSSTRR228 pKa = 11.84RR229 pKa = 11.84DD230 pKa = 3.33SRR232 pKa = 11.84SRR234 pKa = 11.84KK235 pKa = 9.62HH236 pKa = 6.27EE237 pKa = 3.53IAQRR241 pKa = 11.84RR242 pKa = 11.84VCSYY246 pKa = 10.13QKK248 pKa = 10.53HH249 pKa = 5.5SAPTTIKK256 pKa = 10.52PFQRR260 pKa = 11.84WNNILLNEE268 pKa = 4.08LQEE271 pKa = 4.28GAEE274 pKa = 4.11YY275 pKa = 10.84EE276 pKa = 4.01RR277 pKa = 11.84TNRR280 pKa = 11.84KK281 pKa = 9.03RR282 pKa = 11.84SPSIRR287 pKa = 11.84PRR289 pKa = 11.84EE290 pKa = 4.01QSPVRR295 pKa = 11.84HH296 pKa = 5.69NQRR299 pKa = 11.84CSGRR303 pKa = 11.84VLISKK308 pKa = 8.67QANHH312 pKa = 5.75QKK314 pKa = 9.21ATEE317 pKa = 4.21QPCSEE322 pKa = 3.94RR323 pKa = 11.84SAVQKK328 pKa = 10.78RR329 pKa = 11.84GWCKK333 pKa = 10.16LSSLARR339 pKa = 11.84PSLGFCIHH347 pKa = 6.2QGPLAFVTKK356 pKa = 9.89PVYY359 pKa = 10.29FGCTNSPHH367 pKa = 5.07VQQTILYY374 pKa = 8.66KK375 pKa = 10.69LVEE378 pKa = 4.41TVSGLFLTHH387 pKa = 6.93LNFVHH392 pKa = 7.96DD393 pKa = 4.59IGCSRR398 pKa = 11.84KK399 pKa = 10.19SFWPNSNLPKK409 pKa = 10.14KK410 pKa = 9.73YY411 pKa = 10.22NIVISRR417 pKa = 11.84FWARR421 pKa = 11.84HH422 pKa = 3.93CWQFYY427 pKa = 9.29SPCSKK432 pKa = 10.76AILVPFSSKK441 pKa = 10.6FGIVPALEE449 pKa = 4.14LLGGSLFSRR458 pKa = 11.84YY459 pKa = 7.96MRR461 pKa = 11.84DD462 pKa = 3.12LPGSNRR468 pKa = 11.84LLKK471 pKa = 10.46LSHH474 pKa = 6.74RR475 pKa = 11.84GWGPADD481 pKa = 3.27PVAYY485 pKa = 10.13RR486 pKa = 11.84IMTEE490 pKa = 3.74TTRR493 pKa = 11.84PQNDD497 pKa = 3.43TAQDD501 pKa = 3.36RR502 pKa = 11.84QEE504 pKa = 4.77RR505 pKa = 11.84RR506 pKa = 11.84LQTAKK511 pKa = 10.88RR512 pKa = 11.84LLTEE516 pKa = 4.75ADD518 pKa = 2.79TDD520 pKa = 3.83AAVIEE525 pKa = 4.57DD526 pKa = 4.28CQTVLSRR533 pKa = 11.84PGYY536 pKa = 9.53EE537 pKa = 3.94RR538 pKa = 11.84SHH540 pKa = 6.03SQASLDD546 pKa = 3.57ACSALLALAQGDD558 pKa = 4.02EE559 pKa = 4.36STKK562 pKa = 9.56RR563 pKa = 11.84TRR565 pKa = 11.84EE566 pKa = 3.89TKK568 pKa = 9.96QVLTAVSRR576 pKa = 11.84RR577 pKa = 11.84GGDD580 pKa = 3.36CC581 pKa = 3.77
MM1 pKa = 7.23QNTHH5 pKa = 7.18AIEE8 pKa = 3.98QKK10 pKa = 10.44GRR12 pKa = 11.84GIAVALRR19 pKa = 11.84PSPRR23 pKa = 11.84DD24 pKa = 3.17AGAGSGRR31 pKa = 11.84ASAGASVQPVPAPGALPAGHH51 pKa = 6.89PRR53 pKa = 11.84SHH55 pKa = 6.26EE56 pKa = 3.85VRR58 pKa = 11.84HH59 pKa = 5.27RR60 pKa = 11.84ASRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84TRR67 pKa = 11.84SAPAASFRR75 pKa = 11.84LRR77 pKa = 11.84RR78 pKa = 11.84SGPSPGAPGRR88 pKa = 11.84HH89 pKa = 6.15AGLAGGSRR97 pKa = 11.84PYY99 pKa = 10.53SEE101 pKa = 5.45RR102 pKa = 11.84SYY104 pKa = 11.68SCIQPPRR111 pKa = 11.84EE112 pKa = 4.09IVDD115 pKa = 3.32EE116 pKa = 4.2RR117 pKa = 11.84APDD120 pKa = 3.21VWANGPAAFVLRR132 pKa = 11.84ALAPPCEE139 pKa = 4.24CFIGRR144 pKa = 11.84VCEE147 pKa = 4.02FDD149 pKa = 3.94LLGAVVASDD158 pKa = 4.14DD159 pKa = 3.67GHH161 pKa = 7.24GEE163 pKa = 4.0LVRR166 pKa = 11.84VPVVGRR172 pKa = 11.84AFAVDD177 pKa = 4.07AQEE180 pKa = 3.25QWAALRR186 pKa = 11.84EE187 pKa = 4.24QFTGNTFLRR196 pKa = 11.84MCVSRR201 pKa = 11.84ADD203 pKa = 3.71RR204 pKa = 11.84FTSRR208 pKa = 11.84RR209 pKa = 11.84SRR211 pKa = 11.84LTHH214 pKa = 5.8GSQWNRR220 pKa = 11.84RR221 pKa = 11.84AWCSSTRR228 pKa = 11.84RR229 pKa = 11.84DD230 pKa = 3.33SRR232 pKa = 11.84SRR234 pKa = 11.84KK235 pKa = 9.62HH236 pKa = 6.27EE237 pKa = 3.53IAQRR241 pKa = 11.84RR242 pKa = 11.84VCSYY246 pKa = 10.13QKK248 pKa = 10.53HH249 pKa = 5.5SAPTTIKK256 pKa = 10.52PFQRR260 pKa = 11.84WNNILLNEE268 pKa = 4.08LQEE271 pKa = 4.28GAEE274 pKa = 4.11YY275 pKa = 10.84EE276 pKa = 4.01RR277 pKa = 11.84TNRR280 pKa = 11.84KK281 pKa = 9.03RR282 pKa = 11.84SPSIRR287 pKa = 11.84PRR289 pKa = 11.84EE290 pKa = 4.01QSPVRR295 pKa = 11.84HH296 pKa = 5.69NQRR299 pKa = 11.84CSGRR303 pKa = 11.84VLISKK308 pKa = 8.67QANHH312 pKa = 5.75QKK314 pKa = 9.21ATEE317 pKa = 4.21QPCSEE322 pKa = 3.94RR323 pKa = 11.84SAVQKK328 pKa = 10.78RR329 pKa = 11.84GWCKK333 pKa = 10.16LSSLARR339 pKa = 11.84PSLGFCIHH347 pKa = 6.2QGPLAFVTKK356 pKa = 9.89PVYY359 pKa = 10.29FGCTNSPHH367 pKa = 5.07VQQTILYY374 pKa = 8.66KK375 pKa = 10.69LVEE378 pKa = 4.41TVSGLFLTHH387 pKa = 6.93LNFVHH392 pKa = 7.96DD393 pKa = 4.59IGCSRR398 pKa = 11.84KK399 pKa = 10.19SFWPNSNLPKK409 pKa = 10.14KK410 pKa = 9.73YY411 pKa = 10.22NIVISRR417 pKa = 11.84FWARR421 pKa = 11.84HH422 pKa = 3.93CWQFYY427 pKa = 9.29SPCSKK432 pKa = 10.76AILVPFSSKK441 pKa = 10.6FGIVPALEE449 pKa = 4.14LLGGSLFSRR458 pKa = 11.84YY459 pKa = 7.96MRR461 pKa = 11.84DD462 pKa = 3.12LPGSNRR468 pKa = 11.84LLKK471 pKa = 10.46LSHH474 pKa = 6.74RR475 pKa = 11.84GWGPADD481 pKa = 3.27PVAYY485 pKa = 10.13RR486 pKa = 11.84IMTEE490 pKa = 3.74TTRR493 pKa = 11.84PQNDD497 pKa = 3.43TAQDD501 pKa = 3.36RR502 pKa = 11.84QEE504 pKa = 4.77RR505 pKa = 11.84RR506 pKa = 11.84LQTAKK511 pKa = 10.88RR512 pKa = 11.84LLTEE516 pKa = 4.75ADD518 pKa = 2.79TDD520 pKa = 3.83AAVIEE525 pKa = 4.57DD526 pKa = 4.28CQTVLSRR533 pKa = 11.84PGYY536 pKa = 9.53EE537 pKa = 3.94RR538 pKa = 11.84SHH540 pKa = 6.03SQASLDD546 pKa = 3.57ACSALLALAQGDD558 pKa = 4.02EE559 pKa = 4.36STKK562 pKa = 9.56RR563 pKa = 11.84TRR565 pKa = 11.84EE566 pKa = 3.89TKK568 pKa = 9.96QVLTAVSRR576 pKa = 11.84RR577 pKa = 11.84GGDD580 pKa = 3.36CC581 pKa = 3.77
Molecular weight: 64.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1200597 |
30 |
2306 |
283.6 |
30.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.415 ± 0.044 | 0.757 ± 0.012 |
8.326 ± 0.043 | 8.09 ± 0.051 |
3.254 ± 0.027 | 8.278 ± 0.035 |
2.005 ± 0.017 | 4.386 ± 0.029 |
2.001 ± 0.021 | 8.81 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.878 ± 0.017 | 2.586 ± 0.025 |
4.591 ± 0.027 | 3.124 ± 0.023 |
6.114 ± 0.034 | 5.955 ± 0.033 |
6.916 ± 0.037 | 8.648 ± 0.035 |
1.147 ± 0.016 | 2.718 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
