
Lujinxingia litoralis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Bradymonadales; Bradymonadaceae; Lujinxingia
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
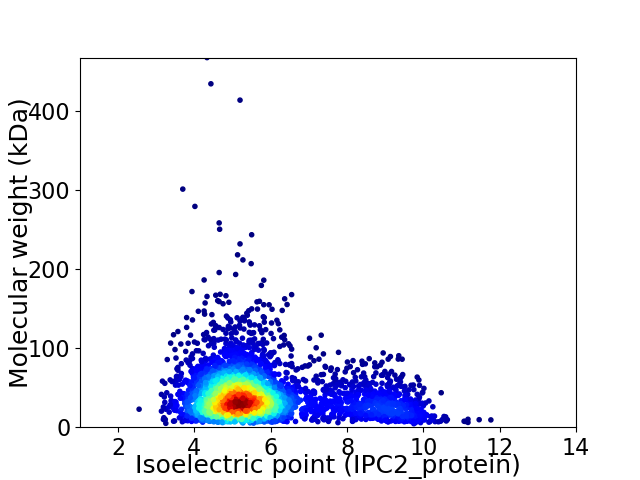
Virtual 2D-PAGE plot for 3776 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328CBD2|A0A328CBD2_9DELT Uncharacterized protein OS=Lujinxingia litoralis OX=2211119 GN=DL240_09360 PE=4 SV=1
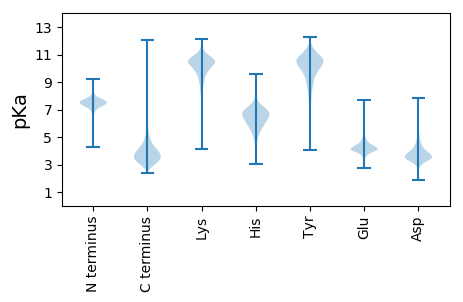
MM1 pKa = 7.56ALSACGGDD9 pKa = 3.88PEE11 pKa = 4.97PDD13 pKa = 3.25RR14 pKa = 11.84NPDD17 pKa = 2.86VGQDD21 pKa = 3.4VDD23 pKa = 4.34PDD25 pKa = 3.95PDD27 pKa = 5.43AGDD30 pKa = 3.63TDD32 pKa = 3.89VDD34 pKa = 3.69PDD36 pKa = 4.25ADD38 pKa = 3.83VDD40 pKa = 3.89PDD42 pKa = 4.15ADD44 pKa = 3.71VDD46 pKa = 3.98PDD48 pKa = 3.76TDD50 pKa = 3.97VDD52 pKa = 3.6ITDD55 pKa = 4.11PPEE58 pKa = 3.95DD59 pKa = 5.42AIACDD64 pKa = 4.08EE65 pKa = 4.82PMPQPPQGEE74 pKa = 4.3RR75 pKa = 11.84CVVIPGNGDD84 pKa = 3.68HH85 pKa = 6.95ILFRR89 pKa = 11.84GTLLAGDD96 pKa = 4.3DD97 pKa = 4.25VYY99 pKa = 11.45HH100 pKa = 6.81EE101 pKa = 5.02GSLLLNDD108 pKa = 3.78QSPNRR113 pKa = 11.84QIVCSGCGCADD124 pKa = 3.42TPEE127 pKa = 4.56AQDD130 pKa = 3.57ATIVSCPSGVISPGLINPHH149 pKa = 5.7DD150 pKa = 5.38HH151 pKa = 5.8ITYY154 pKa = 10.02SLSHH158 pKa = 6.54PRR160 pKa = 11.84PHH162 pKa = 7.58GEE164 pKa = 3.44EE165 pKa = 4.32RR166 pKa = 11.84FDD168 pKa = 4.93HH169 pKa = 5.93RR170 pKa = 11.84HH171 pKa = 4.86DD172 pKa = 3.04WRR174 pKa = 11.84RR175 pKa = 11.84GLRR178 pKa = 11.84GHH180 pKa = 6.48DD181 pKa = 4.25QINTSPGSDD190 pKa = 3.19NSHH193 pKa = 6.26EE194 pKa = 4.67GILYY198 pKa = 10.71GEE200 pKa = 4.29LRR202 pKa = 11.84MLFGGATSVVGSVGSGDD219 pKa = 3.37ASGMLRR225 pKa = 11.84NLDD228 pKa = 3.4NTSYY232 pKa = 11.02TEE234 pKa = 4.05GLSGVDD240 pKa = 2.75VSYY243 pKa = 9.28RR244 pKa = 11.84TFPLGDD250 pKa = 3.47SNGTLRR256 pKa = 11.84ASGCDD261 pKa = 3.45YY262 pKa = 11.26PNIDD266 pKa = 3.51NEE268 pKa = 4.23SRR270 pKa = 11.84LNSGVYY276 pKa = 9.92LPHH279 pKa = 8.02LSEE282 pKa = 6.05GIDD285 pKa = 3.23PEE287 pKa = 4.45ANNEE291 pKa = 3.91FHH293 pKa = 6.87CASGASGSDD302 pKa = 3.92LIQDD306 pKa = 3.61NTSIIHH312 pKa = 6.9GIGLSTRR319 pKa = 11.84DD320 pKa = 3.25IALMARR326 pKa = 11.84RR327 pKa = 11.84GATLVWSARR336 pKa = 11.84TNIDD340 pKa = 3.45LYY342 pKa = 11.39GNTAQAPIFKK352 pKa = 10.38RR353 pKa = 11.84FGVPIALGTDD363 pKa = 3.55WSASGSMNMLRR374 pKa = 11.84EE375 pKa = 4.21LQCADD380 pKa = 3.42YY381 pKa = 11.18LNRR384 pKa = 11.84LYY386 pKa = 10.99YY387 pKa = 10.64DD388 pKa = 3.3EE389 pKa = 5.29TFTEE393 pKa = 4.15QEE395 pKa = 3.9LWMMATANAADD406 pKa = 3.63AMGAGDD412 pKa = 3.52QIGRR416 pKa = 11.84LEE418 pKa = 4.11EE419 pKa = 5.08GYY421 pKa = 11.14VGDD424 pKa = 3.48ITIFDD429 pKa = 3.77GTDD432 pKa = 2.97RR433 pKa = 11.84LPYY436 pKa = 9.9RR437 pKa = 11.84AIIDD441 pKa = 3.9AEE443 pKa = 3.92IADD446 pKa = 3.72IVLVLRR452 pKa = 11.84GGEE455 pKa = 4.14PLYY458 pKa = 11.38GDD460 pKa = 3.72AQLIEE465 pKa = 4.47ALVDD469 pKa = 3.62SAEE472 pKa = 4.61LDD474 pKa = 3.45GCEE477 pKa = 4.66QIDD480 pKa = 3.58VCEE483 pKa = 4.16RR484 pKa = 11.84GRR486 pKa = 11.84RR487 pKa = 11.84LCVEE491 pKa = 4.28LDD493 pKa = 3.18AGKK496 pKa = 10.13SLSAIRR502 pKa = 11.84SAVSSNAYY510 pKa = 9.65EE511 pKa = 4.44LFFCGEE517 pKa = 4.1PDD519 pKa = 4.97DD520 pKa = 5.52EE521 pKa = 4.7PSCMPFRR528 pKa = 11.84PNEE531 pKa = 4.0YY532 pKa = 10.41SGLTDD537 pKa = 3.34NTDD540 pKa = 2.99NSGDD544 pKa = 4.16GIPDD548 pKa = 3.51AVDD551 pKa = 3.02NCPAYY556 pKa = 10.42FNPIRR561 pKa = 11.84PMDD564 pKa = 4.33GGQQPDD570 pKa = 3.61TNGNGIGDD578 pKa = 3.74ICDD581 pKa = 3.91PCPLSEE587 pKa = 5.77DD588 pKa = 4.32PNCNTIDD595 pKa = 4.49PDD597 pKa = 4.22DD598 pKa = 5.08LDD600 pKa = 5.86GDD602 pKa = 4.38GVANDD607 pKa = 3.83TDD609 pKa = 3.53NCPVHH614 pKa = 6.71FNPGQEE620 pKa = 4.13NTSGDD625 pKa = 3.99AYY627 pKa = 10.89GDD629 pKa = 3.59ACSPCPEE636 pKa = 4.18TFLGEE641 pKa = 4.5GEE643 pKa = 4.32ACPVSIYY650 pKa = 10.32SIKK653 pKa = 11.01NGTTDD658 pKa = 3.72PGSLGTFEE666 pKa = 4.79GVIVTAVAEE675 pKa = 4.43GEE677 pKa = 4.55GFFVQVDD684 pKa = 4.06PQSDD688 pKa = 4.12DD689 pKa = 3.62YY690 pKa = 11.9QGDD693 pKa = 3.95QYY695 pKa = 11.86SGIYY699 pKa = 9.32VYY701 pKa = 11.07NRR703 pKa = 11.84GGTVFPQVGDD713 pKa = 4.15RR714 pKa = 11.84IDD716 pKa = 3.59LTGSSTLFYY725 pKa = 11.08GQFQVGNVSAINILEE740 pKa = 4.14SGYY743 pKa = 9.54PLPAPTVVSPAEE755 pKa = 3.99VANNGALRR763 pKa = 11.84QAYY766 pKa = 9.16EE767 pKa = 4.18GVLVRR772 pKa = 11.84VEE774 pKa = 4.65DD775 pKa = 3.61VTVTNNSPDD784 pKa = 3.9PGPGQGDD791 pKa = 3.64NPFEE795 pKa = 4.1FAVDD799 pKa = 3.01SGLRR803 pKa = 11.84INNLMYY809 pKa = 10.52TIDD812 pKa = 4.21PKK814 pKa = 11.26PEE816 pKa = 3.87VGNSFASITGVLRR829 pKa = 11.84WANEE833 pKa = 3.73NSKK836 pKa = 10.37VEE838 pKa = 4.11PRR840 pKa = 11.84SEE842 pKa = 4.33LDD844 pKa = 3.62VVSGPPFLAAASPEE858 pKa = 3.96ALFIDD863 pKa = 4.4ADD865 pKa = 4.01GADD868 pKa = 3.93GQLTLSLNRR877 pKa = 11.84ASQGEE882 pKa = 4.43STLALSYY889 pKa = 10.83NPAGIISGPTSATLADD905 pKa = 4.34GEE907 pKa = 4.39QSVTVAIAATTPDD920 pKa = 3.49AEE922 pKa = 4.36ATISVTLDD930 pKa = 3.01GVTLTIPVTTYY941 pKa = 10.25SAASPRR947 pKa = 11.84EE948 pKa = 3.99LSSLSASADD957 pKa = 3.7TIFVGDD963 pKa = 3.62QVNFDD968 pKa = 4.59LEE970 pKa = 4.62LNLPAGAAGEE980 pKa = 4.42TVSLNLLPVEE990 pKa = 4.38TTLPFPAEE998 pKa = 3.84VSFAAGEE1005 pKa = 3.86QRR1007 pKa = 11.84ANITLTFNEE1016 pKa = 4.58GAGDD1020 pKa = 3.81YY1021 pKa = 9.95TLEE1024 pKa = 4.31ATLGTTTLSADD1035 pKa = 3.52VTVANAPDD1043 pKa = 3.65EE1044 pKa = 4.11QSEE1047 pKa = 4.61SFINFDD1053 pKa = 4.44GPGNTYY1059 pKa = 10.61GAGSFVGDD1067 pKa = 3.05SGYY1070 pKa = 9.27TFNYY1074 pKa = 8.0TGGRR1078 pKa = 11.84LVDD1081 pKa = 3.49EE1082 pKa = 5.11TSNSDD1087 pKa = 3.27YY1088 pKa = 11.05TIDD1091 pKa = 4.12GRR1093 pKa = 11.84GLMFGGSGDD1102 pKa = 3.26KK1103 pKa = 10.92SLIVQGLEE1111 pKa = 3.74GGINSLRR1118 pKa = 11.84LEE1120 pKa = 3.86MRR1122 pKa = 11.84KK1123 pKa = 9.9AFTSGANRR1131 pKa = 11.84QIEE1134 pKa = 4.46VFVNGTSVGTSEE1146 pKa = 4.55VFGNASGADD1155 pKa = 3.59ATVHH1159 pKa = 6.16EE1160 pKa = 5.19LLLEE1164 pKa = 5.0DD1165 pKa = 4.25INISGTFDD1173 pKa = 3.57LEE1175 pKa = 3.81IRR1177 pKa = 11.84SIQSGQVTIDD1187 pKa = 3.36NLVWGSFLPP1196 pKa = 4.17
MM1 pKa = 7.56ALSACGGDD9 pKa = 3.88PEE11 pKa = 4.97PDD13 pKa = 3.25RR14 pKa = 11.84NPDD17 pKa = 2.86VGQDD21 pKa = 3.4VDD23 pKa = 4.34PDD25 pKa = 3.95PDD27 pKa = 5.43AGDD30 pKa = 3.63TDD32 pKa = 3.89VDD34 pKa = 3.69PDD36 pKa = 4.25ADD38 pKa = 3.83VDD40 pKa = 3.89PDD42 pKa = 4.15ADD44 pKa = 3.71VDD46 pKa = 3.98PDD48 pKa = 3.76TDD50 pKa = 3.97VDD52 pKa = 3.6ITDD55 pKa = 4.11PPEE58 pKa = 3.95DD59 pKa = 5.42AIACDD64 pKa = 4.08EE65 pKa = 4.82PMPQPPQGEE74 pKa = 4.3RR75 pKa = 11.84CVVIPGNGDD84 pKa = 3.68HH85 pKa = 6.95ILFRR89 pKa = 11.84GTLLAGDD96 pKa = 4.3DD97 pKa = 4.25VYY99 pKa = 11.45HH100 pKa = 6.81EE101 pKa = 5.02GSLLLNDD108 pKa = 3.78QSPNRR113 pKa = 11.84QIVCSGCGCADD124 pKa = 3.42TPEE127 pKa = 4.56AQDD130 pKa = 3.57ATIVSCPSGVISPGLINPHH149 pKa = 5.7DD150 pKa = 5.38HH151 pKa = 5.8ITYY154 pKa = 10.02SLSHH158 pKa = 6.54PRR160 pKa = 11.84PHH162 pKa = 7.58GEE164 pKa = 3.44EE165 pKa = 4.32RR166 pKa = 11.84FDD168 pKa = 4.93HH169 pKa = 5.93RR170 pKa = 11.84HH171 pKa = 4.86DD172 pKa = 3.04WRR174 pKa = 11.84RR175 pKa = 11.84GLRR178 pKa = 11.84GHH180 pKa = 6.48DD181 pKa = 4.25QINTSPGSDD190 pKa = 3.19NSHH193 pKa = 6.26EE194 pKa = 4.67GILYY198 pKa = 10.71GEE200 pKa = 4.29LRR202 pKa = 11.84MLFGGATSVVGSVGSGDD219 pKa = 3.37ASGMLRR225 pKa = 11.84NLDD228 pKa = 3.4NTSYY232 pKa = 11.02TEE234 pKa = 4.05GLSGVDD240 pKa = 2.75VSYY243 pKa = 9.28RR244 pKa = 11.84TFPLGDD250 pKa = 3.47SNGTLRR256 pKa = 11.84ASGCDD261 pKa = 3.45YY262 pKa = 11.26PNIDD266 pKa = 3.51NEE268 pKa = 4.23SRR270 pKa = 11.84LNSGVYY276 pKa = 9.92LPHH279 pKa = 8.02LSEE282 pKa = 6.05GIDD285 pKa = 3.23PEE287 pKa = 4.45ANNEE291 pKa = 3.91FHH293 pKa = 6.87CASGASGSDD302 pKa = 3.92LIQDD306 pKa = 3.61NTSIIHH312 pKa = 6.9GIGLSTRR319 pKa = 11.84DD320 pKa = 3.25IALMARR326 pKa = 11.84RR327 pKa = 11.84GATLVWSARR336 pKa = 11.84TNIDD340 pKa = 3.45LYY342 pKa = 11.39GNTAQAPIFKK352 pKa = 10.38RR353 pKa = 11.84FGVPIALGTDD363 pKa = 3.55WSASGSMNMLRR374 pKa = 11.84EE375 pKa = 4.21LQCADD380 pKa = 3.42YY381 pKa = 11.18LNRR384 pKa = 11.84LYY386 pKa = 10.99YY387 pKa = 10.64DD388 pKa = 3.3EE389 pKa = 5.29TFTEE393 pKa = 4.15QEE395 pKa = 3.9LWMMATANAADD406 pKa = 3.63AMGAGDD412 pKa = 3.52QIGRR416 pKa = 11.84LEE418 pKa = 4.11EE419 pKa = 5.08GYY421 pKa = 11.14VGDD424 pKa = 3.48ITIFDD429 pKa = 3.77GTDD432 pKa = 2.97RR433 pKa = 11.84LPYY436 pKa = 9.9RR437 pKa = 11.84AIIDD441 pKa = 3.9AEE443 pKa = 3.92IADD446 pKa = 3.72IVLVLRR452 pKa = 11.84GGEE455 pKa = 4.14PLYY458 pKa = 11.38GDD460 pKa = 3.72AQLIEE465 pKa = 4.47ALVDD469 pKa = 3.62SAEE472 pKa = 4.61LDD474 pKa = 3.45GCEE477 pKa = 4.66QIDD480 pKa = 3.58VCEE483 pKa = 4.16RR484 pKa = 11.84GRR486 pKa = 11.84RR487 pKa = 11.84LCVEE491 pKa = 4.28LDD493 pKa = 3.18AGKK496 pKa = 10.13SLSAIRR502 pKa = 11.84SAVSSNAYY510 pKa = 9.65EE511 pKa = 4.44LFFCGEE517 pKa = 4.1PDD519 pKa = 4.97DD520 pKa = 5.52EE521 pKa = 4.7PSCMPFRR528 pKa = 11.84PNEE531 pKa = 4.0YY532 pKa = 10.41SGLTDD537 pKa = 3.34NTDD540 pKa = 2.99NSGDD544 pKa = 4.16GIPDD548 pKa = 3.51AVDD551 pKa = 3.02NCPAYY556 pKa = 10.42FNPIRR561 pKa = 11.84PMDD564 pKa = 4.33GGQQPDD570 pKa = 3.61TNGNGIGDD578 pKa = 3.74ICDD581 pKa = 3.91PCPLSEE587 pKa = 5.77DD588 pKa = 4.32PNCNTIDD595 pKa = 4.49PDD597 pKa = 4.22DD598 pKa = 5.08LDD600 pKa = 5.86GDD602 pKa = 4.38GVANDD607 pKa = 3.83TDD609 pKa = 3.53NCPVHH614 pKa = 6.71FNPGQEE620 pKa = 4.13NTSGDD625 pKa = 3.99AYY627 pKa = 10.89GDD629 pKa = 3.59ACSPCPEE636 pKa = 4.18TFLGEE641 pKa = 4.5GEE643 pKa = 4.32ACPVSIYY650 pKa = 10.32SIKK653 pKa = 11.01NGTTDD658 pKa = 3.72PGSLGTFEE666 pKa = 4.79GVIVTAVAEE675 pKa = 4.43GEE677 pKa = 4.55GFFVQVDD684 pKa = 4.06PQSDD688 pKa = 4.12DD689 pKa = 3.62YY690 pKa = 11.9QGDD693 pKa = 3.95QYY695 pKa = 11.86SGIYY699 pKa = 9.32VYY701 pKa = 11.07NRR703 pKa = 11.84GGTVFPQVGDD713 pKa = 4.15RR714 pKa = 11.84IDD716 pKa = 3.59LTGSSTLFYY725 pKa = 11.08GQFQVGNVSAINILEE740 pKa = 4.14SGYY743 pKa = 9.54PLPAPTVVSPAEE755 pKa = 3.99VANNGALRR763 pKa = 11.84QAYY766 pKa = 9.16EE767 pKa = 4.18GVLVRR772 pKa = 11.84VEE774 pKa = 4.65DD775 pKa = 3.61VTVTNNSPDD784 pKa = 3.9PGPGQGDD791 pKa = 3.64NPFEE795 pKa = 4.1FAVDD799 pKa = 3.01SGLRR803 pKa = 11.84INNLMYY809 pKa = 10.52TIDD812 pKa = 4.21PKK814 pKa = 11.26PEE816 pKa = 3.87VGNSFASITGVLRR829 pKa = 11.84WANEE833 pKa = 3.73NSKK836 pKa = 10.37VEE838 pKa = 4.11PRR840 pKa = 11.84SEE842 pKa = 4.33LDD844 pKa = 3.62VVSGPPFLAAASPEE858 pKa = 3.96ALFIDD863 pKa = 4.4ADD865 pKa = 4.01GADD868 pKa = 3.93GQLTLSLNRR877 pKa = 11.84ASQGEE882 pKa = 4.43STLALSYY889 pKa = 10.83NPAGIISGPTSATLADD905 pKa = 4.34GEE907 pKa = 4.39QSVTVAIAATTPDD920 pKa = 3.49AEE922 pKa = 4.36ATISVTLDD930 pKa = 3.01GVTLTIPVTTYY941 pKa = 10.25SAASPRR947 pKa = 11.84EE948 pKa = 3.99LSSLSASADD957 pKa = 3.7TIFVGDD963 pKa = 3.62QVNFDD968 pKa = 4.59LEE970 pKa = 4.62LNLPAGAAGEE980 pKa = 4.42TVSLNLLPVEE990 pKa = 4.38TTLPFPAEE998 pKa = 3.84VSFAAGEE1005 pKa = 3.86QRR1007 pKa = 11.84ANITLTFNEE1016 pKa = 4.58GAGDD1020 pKa = 3.81YY1021 pKa = 9.95TLEE1024 pKa = 4.31ATLGTTTLSADD1035 pKa = 3.52VTVANAPDD1043 pKa = 3.65EE1044 pKa = 4.11QSEE1047 pKa = 4.61SFINFDD1053 pKa = 4.44GPGNTYY1059 pKa = 10.61GAGSFVGDD1067 pKa = 3.05SGYY1070 pKa = 9.27TFNYY1074 pKa = 8.0TGGRR1078 pKa = 11.84LVDD1081 pKa = 3.49EE1082 pKa = 5.11TSNSDD1087 pKa = 3.27YY1088 pKa = 11.05TIDD1091 pKa = 4.12GRR1093 pKa = 11.84GLMFGGSGDD1102 pKa = 3.26KK1103 pKa = 10.92SLIVQGLEE1111 pKa = 3.74GGINSLRR1118 pKa = 11.84LEE1120 pKa = 3.86MRR1122 pKa = 11.84KK1123 pKa = 9.9AFTSGANRR1131 pKa = 11.84QIEE1134 pKa = 4.46VFVNGTSVGTSEE1146 pKa = 4.55VFGNASGADD1155 pKa = 3.59ATVHH1159 pKa = 6.16EE1160 pKa = 5.19LLLEE1164 pKa = 5.0DD1165 pKa = 4.25INISGTFDD1173 pKa = 3.57LEE1175 pKa = 3.81IRR1177 pKa = 11.84SIQSGQVTIDD1187 pKa = 3.36NLVWGSFLPP1196 pKa = 4.17
Molecular weight: 126.12 kDa
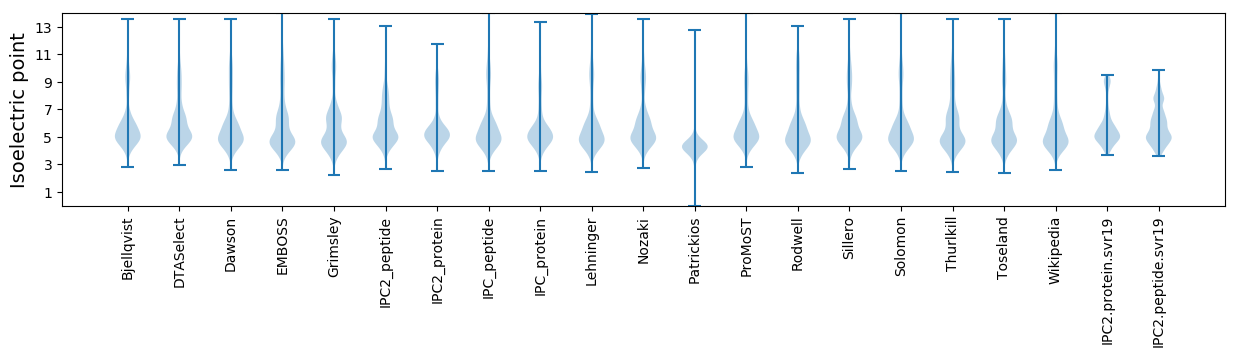
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328C748|A0A328C748_9DELT Peptidase_M23 domain-containing protein OS=Lujinxingia litoralis OX=2211119 GN=DL240_09425 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84SGRR5 pKa = 11.84RR6 pKa = 11.84VRR8 pKa = 11.84GRR10 pKa = 11.84GRR12 pKa = 11.84GRR14 pKa = 11.84ARR16 pKa = 11.84VRR18 pKa = 11.84GRR20 pKa = 11.84VRR22 pKa = 11.84GRR24 pKa = 11.84GRR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84VRR32 pKa = 11.84ARR34 pKa = 11.84ARR36 pKa = 11.84ARR38 pKa = 11.84ARR40 pKa = 11.84GRR42 pKa = 11.84VRR44 pKa = 11.84VRR46 pKa = 11.84GRR48 pKa = 11.84GRR50 pKa = 11.84GRR52 pKa = 11.84GRR54 pKa = 11.84GRR56 pKa = 11.84GRR58 pKa = 11.84GRR60 pKa = 11.84VRR62 pKa = 11.84GRR64 pKa = 11.84VRR66 pKa = 11.84ARR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 7.79TPHH73 pKa = 6.15LQRR76 pKa = 11.84LPGVGCFF83 pKa = 3.82
MM1 pKa = 7.74RR2 pKa = 11.84SGRR5 pKa = 11.84RR6 pKa = 11.84VRR8 pKa = 11.84GRR10 pKa = 11.84GRR12 pKa = 11.84GRR14 pKa = 11.84ARR16 pKa = 11.84VRR18 pKa = 11.84GRR20 pKa = 11.84VRR22 pKa = 11.84GRR24 pKa = 11.84GRR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84VRR32 pKa = 11.84ARR34 pKa = 11.84ARR36 pKa = 11.84ARR38 pKa = 11.84ARR40 pKa = 11.84GRR42 pKa = 11.84VRR44 pKa = 11.84VRR46 pKa = 11.84GRR48 pKa = 11.84GRR50 pKa = 11.84GRR52 pKa = 11.84GRR54 pKa = 11.84GRR56 pKa = 11.84GRR58 pKa = 11.84GRR60 pKa = 11.84VRR62 pKa = 11.84GRR64 pKa = 11.84VRR66 pKa = 11.84ARR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 7.79TPHH73 pKa = 6.15LQRR76 pKa = 11.84LPGVGCFF83 pKa = 3.82
Molecular weight: 9.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1447238 |
37 |
4109 |
383.3 |
41.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.323 ± 0.058 | 1.131 ± 0.034 |
5.85 ± 0.038 | 7.575 ± 0.049 |
3.445 ± 0.024 | 8.221 ± 0.043 |
2.303 ± 0.021 | 4.119 ± 0.029 |
2.24 ± 0.026 | 10.653 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.111 ± 0.017 | 2.377 ± 0.025 |
5.441 ± 0.037 | 3.746 ± 0.025 |
7.703 ± 0.047 | 5.71 ± 0.029 |
5.047 ± 0.034 | 7.343 ± 0.038 |
1.313 ± 0.017 | 2.348 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
