
Torque teno sus virus SH0822/2008
Taxonomy: Viruses; Anelloviridae; Iotatorquevirus; unclassified Iotatorquevirus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
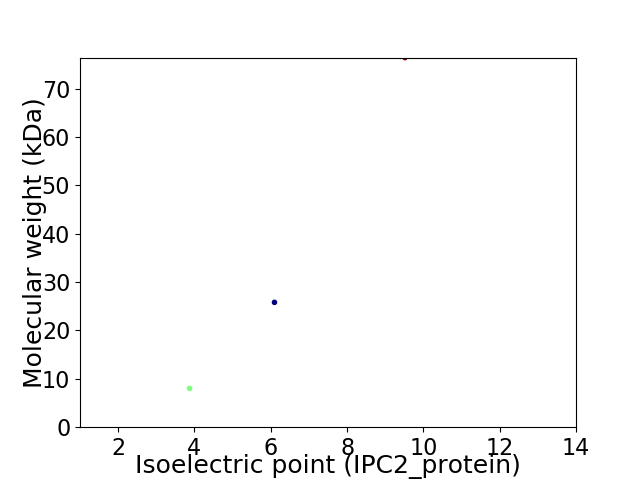
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5K6A1|D5K6A1_9VIRU Capsid protein OS=Torque teno sus virus SH0822/2008 OX=742012 PE=3 SV=1
MM1 pKa = 7.59PEE3 pKa = 3.57HH4 pKa = 6.88WEE6 pKa = 4.15EE7 pKa = 3.82AWLEE11 pKa = 4.1ATKK14 pKa = 10.4GWHH17 pKa = 7.41DD18 pKa = 4.1LDD20 pKa = 4.34CRR22 pKa = 11.84CGNWQDD28 pKa = 4.29HH29 pKa = 5.86LWLLLADD36 pKa = 4.62GDD38 pKa = 4.33AALAAAVDD46 pKa = 4.65AIEE49 pKa = 4.59RR50 pKa = 11.84DD51 pKa = 3.58AMDD54 pKa = 5.12GEE56 pKa = 4.83DD57 pKa = 3.73ATTATDD63 pKa = 3.32RR64 pKa = 11.84VTIGDD69 pKa = 4.33DD70 pKa = 3.11GWW72 pKa = 5.36
MM1 pKa = 7.59PEE3 pKa = 3.57HH4 pKa = 6.88WEE6 pKa = 4.15EE7 pKa = 3.82AWLEE11 pKa = 4.1ATKK14 pKa = 10.4GWHH17 pKa = 7.41DD18 pKa = 4.1LDD20 pKa = 4.34CRR22 pKa = 11.84CGNWQDD28 pKa = 4.29HH29 pKa = 5.86LWLLLADD36 pKa = 4.62GDD38 pKa = 4.33AALAAAVDD46 pKa = 4.65AIEE49 pKa = 4.59RR50 pKa = 11.84DD51 pKa = 3.58AMDD54 pKa = 5.12GEE56 pKa = 4.83DD57 pKa = 3.73ATTATDD63 pKa = 3.32RR64 pKa = 11.84VTIGDD69 pKa = 4.33DD70 pKa = 3.11GWW72 pKa = 5.36
Molecular weight: 8.02 kDa
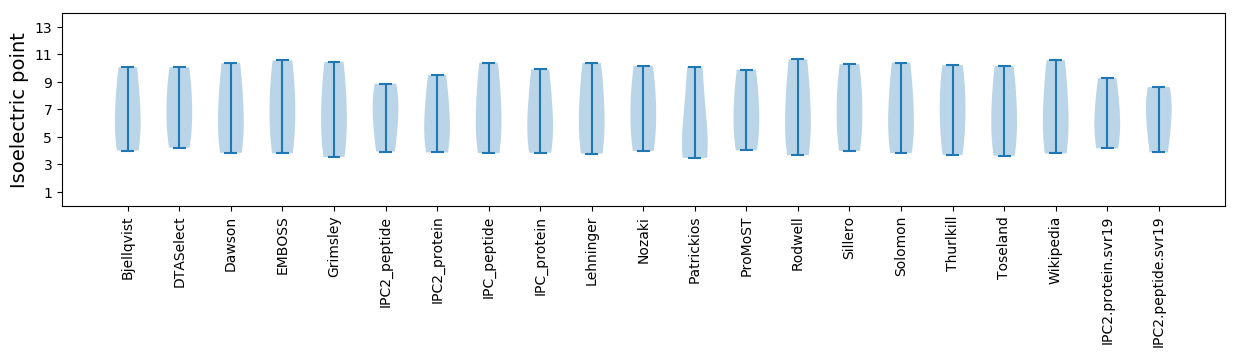
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5K6A1|D5K6A1_9VIRU Capsid protein OS=Torque teno sus virus SH0822/2008 OX=742012 PE=3 SV=1
MM1 pKa = 7.55AFARR5 pKa = 11.84RR6 pKa = 11.84WRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84FGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 8.4RR19 pKa = 11.84KK20 pKa = 9.16RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 7.49GWRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.05YY30 pKa = 9.6RR31 pKa = 11.84YY32 pKa = 9.48RR33 pKa = 11.84PRR35 pKa = 11.84YY36 pKa = 7.87YY37 pKa = 9.67RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84WLVRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SVYY51 pKa = 9.57RR52 pKa = 11.84RR53 pKa = 11.84GGRR56 pKa = 11.84RR57 pKa = 11.84ARR59 pKa = 11.84PYY61 pKa = 10.18RR62 pKa = 11.84ISAFNPKK69 pKa = 9.02VMRR72 pKa = 11.84KK73 pKa = 8.14VVIRR77 pKa = 11.84GWWPILQCLKK87 pKa = 10.27GQEE90 pKa = 4.22SLRR93 pKa = 11.84YY94 pKa = 9.4RR95 pKa = 11.84PLQWDD100 pKa = 3.67TEE102 pKa = 4.37KK103 pKa = 10.53QWRR106 pKa = 11.84VKK108 pKa = 10.78ADD110 pKa = 3.24YY111 pKa = 10.03EE112 pKa = 4.44DD113 pKa = 3.77NYY115 pKa = 11.1GYY117 pKa = 10.8LVQYY121 pKa = 10.35GGGWGSGEE129 pKa = 3.98VTLEE133 pKa = 3.75GLYY136 pKa = 9.79QEE138 pKa = 4.53HH139 pKa = 6.78LLWRR143 pKa = 11.84NSWSKK148 pKa = 11.58GNDD151 pKa = 2.87GMDD154 pKa = 3.61LVRR157 pKa = 11.84YY158 pKa = 7.43FGCIVYY164 pKa = 9.63LYY166 pKa = 9.39PLQDD170 pKa = 2.88QDD172 pKa = 3.81YY173 pKa = 8.02WFWWDD178 pKa = 3.03TDD180 pKa = 3.83FKK182 pKa = 10.92EE183 pKa = 5.59LYY185 pKa = 10.35AEE187 pKa = 4.46SIKK190 pKa = 10.6EE191 pKa = 3.89YY192 pKa = 9.85SQPSVMMMAKK202 pKa = 8.79RR203 pKa = 11.84TKK205 pKa = 10.53LVIARR210 pKa = 11.84DD211 pKa = 3.8RR212 pKa = 11.84APHH215 pKa = 5.55RR216 pKa = 11.84RR217 pKa = 11.84KK218 pKa = 9.54VRR220 pKa = 11.84KK221 pKa = 9.5IFIPPPSRR229 pKa = 11.84DD230 pKa = 3.08TTQWQFQTDD239 pKa = 3.92FCKK242 pKa = 10.63RR243 pKa = 11.84PLFTWAAGLIDD254 pKa = 3.75MQKK257 pKa = 10.58PFDD260 pKa = 4.05ANGAFRR266 pKa = 11.84NAWWLEE272 pKa = 3.59TRR274 pKa = 11.84NDD276 pKa = 3.67QGEE279 pKa = 4.24MKK281 pKa = 10.59YY282 pKa = 10.05IEE284 pKa = 4.17LWGRR288 pKa = 11.84VPPQGDD294 pKa = 3.53TEE296 pKa = 4.47LPKK299 pKa = 10.82QSDD302 pKa = 3.78FKK304 pKa = 11.38KK305 pKa = 10.9GDD307 pKa = 3.29NNPNYY312 pKa = 10.16NIQEE316 pKa = 4.02GHH318 pKa = 5.77EE319 pKa = 4.15KK320 pKa = 10.17KK321 pKa = 10.11IYY323 pKa = 9.71PIIIYY328 pKa = 10.07VDD330 pKa = 3.23QKK332 pKa = 10.86DD333 pKa = 3.27QKK335 pKa = 8.94TRR337 pKa = 11.84KK338 pKa = 9.19KK339 pKa = 11.05YY340 pKa = 9.24CVCYY344 pKa = 10.77NKK346 pKa = 9.49TLNRR350 pKa = 11.84WRR352 pKa = 11.84KK353 pKa = 8.39AQASTLAIGDD363 pKa = 3.73LQGLVLRR370 pKa = 11.84QLMNQEE376 pKa = 3.38MTYY379 pKa = 9.14YY380 pKa = 8.94WKK382 pKa = 10.71SGEE385 pKa = 4.0FSSPFMQRR393 pKa = 11.84WKK395 pKa = 9.38GTRR398 pKa = 11.84LITIDD403 pKa = 3.28ARR405 pKa = 11.84RR406 pKa = 11.84ADD408 pKa = 3.79TEE410 pKa = 4.14NPKK413 pKa = 10.01VSSWEE418 pKa = 3.64WGQNWNTNGTVLKK431 pKa = 10.4EE432 pKa = 3.89VFNINLTNTQIRR444 pKa = 11.84QDD446 pKa = 3.96DD447 pKa = 4.08FAKK450 pKa = 9.65LTLPKK455 pKa = 10.03SPHH458 pKa = 6.88DD459 pKa = 3.41IDD461 pKa = 4.84FGHH464 pKa = 7.26HH465 pKa = 5.47SRR467 pKa = 11.84FGPFCVKK474 pKa = 10.61NEE476 pKa = 3.84PLEE479 pKa = 4.21FQLRR483 pKa = 11.84APTPTNLWFQYY494 pKa = 10.74KK495 pKa = 10.14FLFQFGGEE503 pKa = 4.14YY504 pKa = 10.2QPPTGIRR511 pKa = 11.84DD512 pKa = 3.68PCIDD516 pKa = 3.55TPAYY520 pKa = 8.19PVPQSGSVTHH530 pKa = 6.54PKK532 pKa = 10.14FAGKK536 pKa = 10.61GGMLTEE542 pKa = 4.16TDD544 pKa = 2.55RR545 pKa = 11.84WGITTASSRR554 pKa = 11.84TLSADD559 pKa = 3.45TPTEE563 pKa = 4.03AAQSALLRR571 pKa = 11.84GDD573 pKa = 3.66AEE575 pKa = 4.5KK576 pKa = 10.68KK577 pKa = 10.56GEE579 pKa = 4.18EE580 pKa = 4.3TEE582 pKa = 4.32EE583 pKa = 4.22TASSSSITSAEE594 pKa = 4.02SSTEE598 pKa = 3.92GDD600 pKa = 3.33GSSDD604 pKa = 3.52DD605 pKa = 3.84EE606 pKa = 4.06EE607 pKa = 4.58TIRR610 pKa = 11.84RR611 pKa = 11.84RR612 pKa = 11.84RR613 pKa = 11.84RR614 pKa = 11.84TWKK617 pKa = 9.54RR618 pKa = 11.84LRR620 pKa = 11.84RR621 pKa = 11.84MVRR624 pKa = 11.84EE625 pKa = 3.76QLDD628 pKa = 2.95RR629 pKa = 11.84RR630 pKa = 11.84MDD632 pKa = 3.46HH633 pKa = 6.39KK634 pKa = 10.63RR635 pKa = 11.84QRR637 pKa = 11.84LHH639 pKa = 6.44
MM1 pKa = 7.55AFARR5 pKa = 11.84RR6 pKa = 11.84WRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84FGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 8.4RR19 pKa = 11.84KK20 pKa = 9.16RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 7.49GWRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.05YY30 pKa = 9.6RR31 pKa = 11.84YY32 pKa = 9.48RR33 pKa = 11.84PRR35 pKa = 11.84YY36 pKa = 7.87YY37 pKa = 9.67RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84WLVRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SVYY51 pKa = 9.57RR52 pKa = 11.84RR53 pKa = 11.84GGRR56 pKa = 11.84RR57 pKa = 11.84ARR59 pKa = 11.84PYY61 pKa = 10.18RR62 pKa = 11.84ISAFNPKK69 pKa = 9.02VMRR72 pKa = 11.84KK73 pKa = 8.14VVIRR77 pKa = 11.84GWWPILQCLKK87 pKa = 10.27GQEE90 pKa = 4.22SLRR93 pKa = 11.84YY94 pKa = 9.4RR95 pKa = 11.84PLQWDD100 pKa = 3.67TEE102 pKa = 4.37KK103 pKa = 10.53QWRR106 pKa = 11.84VKK108 pKa = 10.78ADD110 pKa = 3.24YY111 pKa = 10.03EE112 pKa = 4.44DD113 pKa = 3.77NYY115 pKa = 11.1GYY117 pKa = 10.8LVQYY121 pKa = 10.35GGGWGSGEE129 pKa = 3.98VTLEE133 pKa = 3.75GLYY136 pKa = 9.79QEE138 pKa = 4.53HH139 pKa = 6.78LLWRR143 pKa = 11.84NSWSKK148 pKa = 11.58GNDD151 pKa = 2.87GMDD154 pKa = 3.61LVRR157 pKa = 11.84YY158 pKa = 7.43FGCIVYY164 pKa = 9.63LYY166 pKa = 9.39PLQDD170 pKa = 2.88QDD172 pKa = 3.81YY173 pKa = 8.02WFWWDD178 pKa = 3.03TDD180 pKa = 3.83FKK182 pKa = 10.92EE183 pKa = 5.59LYY185 pKa = 10.35AEE187 pKa = 4.46SIKK190 pKa = 10.6EE191 pKa = 3.89YY192 pKa = 9.85SQPSVMMMAKK202 pKa = 8.79RR203 pKa = 11.84TKK205 pKa = 10.53LVIARR210 pKa = 11.84DD211 pKa = 3.8RR212 pKa = 11.84APHH215 pKa = 5.55RR216 pKa = 11.84RR217 pKa = 11.84KK218 pKa = 9.54VRR220 pKa = 11.84KK221 pKa = 9.5IFIPPPSRR229 pKa = 11.84DD230 pKa = 3.08TTQWQFQTDD239 pKa = 3.92FCKK242 pKa = 10.63RR243 pKa = 11.84PLFTWAAGLIDD254 pKa = 3.75MQKK257 pKa = 10.58PFDD260 pKa = 4.05ANGAFRR266 pKa = 11.84NAWWLEE272 pKa = 3.59TRR274 pKa = 11.84NDD276 pKa = 3.67QGEE279 pKa = 4.24MKK281 pKa = 10.59YY282 pKa = 10.05IEE284 pKa = 4.17LWGRR288 pKa = 11.84VPPQGDD294 pKa = 3.53TEE296 pKa = 4.47LPKK299 pKa = 10.82QSDD302 pKa = 3.78FKK304 pKa = 11.38KK305 pKa = 10.9GDD307 pKa = 3.29NNPNYY312 pKa = 10.16NIQEE316 pKa = 4.02GHH318 pKa = 5.77EE319 pKa = 4.15KK320 pKa = 10.17KK321 pKa = 10.11IYY323 pKa = 9.71PIIIYY328 pKa = 10.07VDD330 pKa = 3.23QKK332 pKa = 10.86DD333 pKa = 3.27QKK335 pKa = 8.94TRR337 pKa = 11.84KK338 pKa = 9.19KK339 pKa = 11.05YY340 pKa = 9.24CVCYY344 pKa = 10.77NKK346 pKa = 9.49TLNRR350 pKa = 11.84WRR352 pKa = 11.84KK353 pKa = 8.39AQASTLAIGDD363 pKa = 3.73LQGLVLRR370 pKa = 11.84QLMNQEE376 pKa = 3.38MTYY379 pKa = 9.14YY380 pKa = 8.94WKK382 pKa = 10.71SGEE385 pKa = 4.0FSSPFMQRR393 pKa = 11.84WKK395 pKa = 9.38GTRR398 pKa = 11.84LITIDD403 pKa = 3.28ARR405 pKa = 11.84RR406 pKa = 11.84ADD408 pKa = 3.79TEE410 pKa = 4.14NPKK413 pKa = 10.01VSSWEE418 pKa = 3.64WGQNWNTNGTVLKK431 pKa = 10.4EE432 pKa = 3.89VFNINLTNTQIRR444 pKa = 11.84QDD446 pKa = 3.96DD447 pKa = 4.08FAKK450 pKa = 9.65LTLPKK455 pKa = 10.03SPHH458 pKa = 6.88DD459 pKa = 3.41IDD461 pKa = 4.84FGHH464 pKa = 7.26HH465 pKa = 5.47SRR467 pKa = 11.84FGPFCVKK474 pKa = 10.61NEE476 pKa = 3.84PLEE479 pKa = 4.21FQLRR483 pKa = 11.84APTPTNLWFQYY494 pKa = 10.74KK495 pKa = 10.14FLFQFGGEE503 pKa = 4.14YY504 pKa = 10.2QPPTGIRR511 pKa = 11.84DD512 pKa = 3.68PCIDD516 pKa = 3.55TPAYY520 pKa = 8.19PVPQSGSVTHH530 pKa = 6.54PKK532 pKa = 10.14FAGKK536 pKa = 10.61GGMLTEE542 pKa = 4.16TDD544 pKa = 2.55RR545 pKa = 11.84WGITTASSRR554 pKa = 11.84TLSADD559 pKa = 3.45TPTEE563 pKa = 4.03AAQSALLRR571 pKa = 11.84GDD573 pKa = 3.66AEE575 pKa = 4.5KK576 pKa = 10.68KK577 pKa = 10.56GEE579 pKa = 4.18EE580 pKa = 4.3TEE582 pKa = 4.32EE583 pKa = 4.22TASSSSITSAEE594 pKa = 4.02SSTEE598 pKa = 3.92GDD600 pKa = 3.33GSSDD604 pKa = 3.52DD605 pKa = 3.84EE606 pKa = 4.06EE607 pKa = 4.58TIRR610 pKa = 11.84RR611 pKa = 11.84RR612 pKa = 11.84RR613 pKa = 11.84RR614 pKa = 11.84TWKK617 pKa = 9.54RR618 pKa = 11.84LRR620 pKa = 11.84RR621 pKa = 11.84MVRR624 pKa = 11.84EE625 pKa = 3.76QLDD628 pKa = 2.95RR629 pKa = 11.84RR630 pKa = 11.84MDD632 pKa = 3.46HH633 pKa = 6.39KK634 pKa = 10.63RR635 pKa = 11.84QRR637 pKa = 11.84LHH639 pKa = 6.44
Molecular weight: 76.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
939 |
72 |
639 |
313.0 |
36.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.496 ± 2.476 | 1.597 ± 0.557 |
7.348 ± 2.299 | 6.177 ± 0.804 |
2.982 ± 1.226 | 6.39 ± 0.637 |
2.236 ± 0.917 | 3.834 ± 0.301 |
6.177 ± 0.997 | 7.242 ± 0.803 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.115 | 2.982 ± 0.691 |
5.325 ± 0.85 | 5.005 ± 0.822 |
10.756 ± 1.99 | 5.431 ± 1.21 |
6.177 ± 0.21 | 3.94 ± 0.365 |
4.26 ± 0.869 | 3.408 ± 1.761 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
