
Aminobacter sp. J15
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Aminobacter; unclassified Aminobacter
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
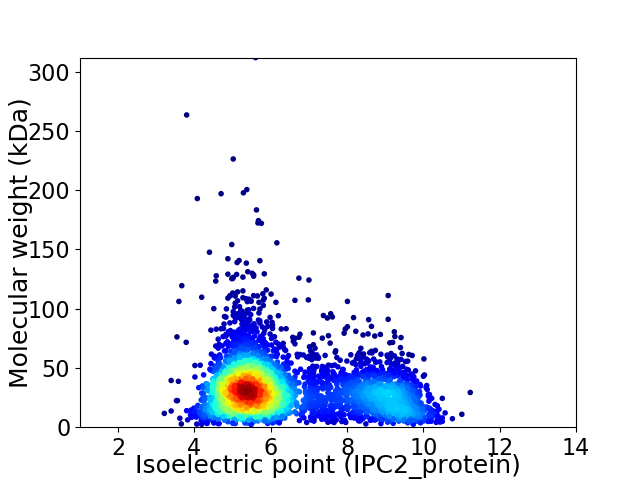
Virtual 2D-PAGE plot for 3999 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A562F9V1|A0A562F9V1_9RHIZ CysZ protein OS=Aminobacter sp. J15 OX=935260 GN=L611_000300000600 PE=4 SV=1
MM1 pKa = 7.54TGMDD5 pKa = 4.55RR6 pKa = 11.84ISEE9 pKa = 4.41EE10 pKa = 3.62IAHH13 pKa = 6.91FIGMFNITVEE23 pKa = 4.15QARR26 pKa = 11.84LRR28 pKa = 11.84DD29 pKa = 3.76AYY31 pKa = 10.93HH32 pKa = 6.78EE33 pKa = 4.54FNPLPAVPSNPEE45 pKa = 3.97LPEE48 pKa = 5.03APASDD53 pKa = 4.46FAAPHH58 pKa = 6.01TLLGYY63 pKa = 10.55DD64 pKa = 3.06PGMRR68 pKa = 11.84YY69 pKa = 9.45APPGWSLEE77 pKa = 4.24EE78 pKa = 3.89VSPWLPLKK86 pKa = 10.66YY87 pKa = 10.09KK88 pKa = 9.07YY89 pKa = 10.89FKK91 pKa = 9.93MQAPGQYY98 pKa = 10.09PFGHH102 pKa = 6.5PQGPSPEE109 pKa = 4.76PGSGMNINITLPEE122 pKa = 4.59IDD124 pKa = 4.4PPGSVASYY132 pKa = 9.86IVQNIYY138 pKa = 10.93LSDD141 pKa = 3.4NDD143 pKa = 3.69YY144 pKa = 11.26FGVGGHH150 pKa = 5.99GLKK153 pKa = 10.73FSPQPIDD160 pKa = 3.96NGDD163 pKa = 4.52LINLALEE170 pKa = 4.46ALSISPIGSHH180 pKa = 5.86EE181 pKa = 4.2MPSGAEE187 pKa = 4.01AIKK190 pKa = 10.91DD191 pKa = 4.33FITSVAEE198 pKa = 3.69EE199 pKa = 4.6LEE201 pKa = 4.39NYY203 pKa = 9.15TPDD206 pKa = 3.25TDD208 pKa = 4.14GPAEE212 pKa = 3.75IHH214 pKa = 5.91VFSGKK219 pKa = 8.11TLDD222 pKa = 4.26GIYY225 pKa = 10.98VNGKK229 pKa = 9.36LVTEE233 pKa = 4.88APNIDD238 pKa = 4.85DD239 pKa = 3.69YY240 pKa = 11.9HH241 pKa = 8.26SFDD244 pKa = 3.91PEE246 pKa = 3.84EE247 pKa = 5.4AEE249 pKa = 4.4EE250 pKa = 4.78GSPEE254 pKa = 3.96HH255 pKa = 7.17KK256 pKa = 9.93PIKK259 pKa = 9.49NVKK262 pKa = 7.71ITYY265 pKa = 8.87DD266 pKa = 2.83GHH268 pKa = 4.93MQIEE272 pKa = 4.38ASVSISAGGNTVVNDD287 pKa = 3.88AVLTNIWTGGTVTVVAGDD305 pKa = 3.85YY306 pKa = 9.75FQLDD310 pKa = 3.4AVIQINAICDD320 pKa = 3.75EE321 pKa = 4.62DD322 pKa = 4.32TLSPTITNWSVDD334 pKa = 3.28DD335 pKa = 3.85ATNQFFNIATFKK347 pKa = 9.57HH348 pKa = 5.72TNPLDD353 pKa = 4.24NNDD356 pKa = 3.71GAHH359 pKa = 6.81LSGSPDD365 pKa = 3.41FPSYY369 pKa = 10.39WIVTEE374 pKa = 3.82ITGDD378 pKa = 3.78LLIVNWLDD386 pKa = 3.3QYY388 pKa = 11.4IFQSDD393 pKa = 3.61NDD395 pKa = 3.73VGIVSSSGVTTEE407 pKa = 5.51IIAGDD412 pKa = 3.76NLAVNTISIMEE423 pKa = 4.57LAYY426 pKa = 10.25AYY428 pKa = 9.97DD429 pKa = 3.99IIVIGGSWYY438 pKa = 10.07DD439 pKa = 3.42ANIIQQLNVLYY450 pKa = 10.92DD451 pKa = 3.5NDD453 pKa = 5.37LIGAVNGFQTTGEE466 pKa = 4.41GSVSTSGNLLWNEE479 pKa = 3.57ALIYY483 pKa = 10.44SVSGDD488 pKa = 3.8YY489 pKa = 11.54GDD491 pKa = 4.5MPAHH495 pKa = 5.2YY496 pKa = 10.11QEE498 pKa = 5.47LADD501 pKa = 4.26NLAAGKK507 pKa = 9.6EE508 pKa = 4.1VLPSGVLSDD517 pKa = 3.9PALAGNEE524 pKa = 4.06ALRR527 pKa = 11.84VLYY530 pKa = 10.43IEE532 pKa = 5.38GDD534 pKa = 4.41LINLNYY540 pKa = 9.51IRR542 pKa = 11.84QTSIVGDD549 pKa = 3.66SDD551 pKa = 4.24QIALAMNEE559 pKa = 3.95FTPRR563 pKa = 11.84ADD565 pKa = 3.94AEE567 pKa = 4.27WTITTGGNALLNHH580 pKa = 6.78AAIVDD585 pKa = 4.42LDD587 pKa = 3.88SLGNIHH593 pKa = 6.87VGGEE597 pKa = 4.14QYY599 pKa = 10.71SQAMLVQAEE608 pKa = 4.85LISDD612 pKa = 3.82KK613 pKa = 11.24PEE615 pKa = 3.84FWAQDD620 pKa = 3.15PDD622 pKa = 3.84ALVNEE627 pKa = 4.75AVAFLDD633 pKa = 5.13DD634 pKa = 5.52SIDD637 pKa = 4.76DD638 pKa = 4.57GPDD641 pKa = 4.28DD642 pKa = 3.88IPAYY646 pKa = 8.74YY647 pKa = 10.28QPDD650 pKa = 3.56NEE652 pKa = 4.49APGGDD657 pKa = 3.95GLQAMLGG664 pKa = 3.68
MM1 pKa = 7.54TGMDD5 pKa = 4.55RR6 pKa = 11.84ISEE9 pKa = 4.41EE10 pKa = 3.62IAHH13 pKa = 6.91FIGMFNITVEE23 pKa = 4.15QARR26 pKa = 11.84LRR28 pKa = 11.84DD29 pKa = 3.76AYY31 pKa = 10.93HH32 pKa = 6.78EE33 pKa = 4.54FNPLPAVPSNPEE45 pKa = 3.97LPEE48 pKa = 5.03APASDD53 pKa = 4.46FAAPHH58 pKa = 6.01TLLGYY63 pKa = 10.55DD64 pKa = 3.06PGMRR68 pKa = 11.84YY69 pKa = 9.45APPGWSLEE77 pKa = 4.24EE78 pKa = 3.89VSPWLPLKK86 pKa = 10.66YY87 pKa = 10.09KK88 pKa = 9.07YY89 pKa = 10.89FKK91 pKa = 9.93MQAPGQYY98 pKa = 10.09PFGHH102 pKa = 6.5PQGPSPEE109 pKa = 4.76PGSGMNINITLPEE122 pKa = 4.59IDD124 pKa = 4.4PPGSVASYY132 pKa = 9.86IVQNIYY138 pKa = 10.93LSDD141 pKa = 3.4NDD143 pKa = 3.69YY144 pKa = 11.26FGVGGHH150 pKa = 5.99GLKK153 pKa = 10.73FSPQPIDD160 pKa = 3.96NGDD163 pKa = 4.52LINLALEE170 pKa = 4.46ALSISPIGSHH180 pKa = 5.86EE181 pKa = 4.2MPSGAEE187 pKa = 4.01AIKK190 pKa = 10.91DD191 pKa = 4.33FITSVAEE198 pKa = 3.69EE199 pKa = 4.6LEE201 pKa = 4.39NYY203 pKa = 9.15TPDD206 pKa = 3.25TDD208 pKa = 4.14GPAEE212 pKa = 3.75IHH214 pKa = 5.91VFSGKK219 pKa = 8.11TLDD222 pKa = 4.26GIYY225 pKa = 10.98VNGKK229 pKa = 9.36LVTEE233 pKa = 4.88APNIDD238 pKa = 4.85DD239 pKa = 3.69YY240 pKa = 11.9HH241 pKa = 8.26SFDD244 pKa = 3.91PEE246 pKa = 3.84EE247 pKa = 5.4AEE249 pKa = 4.4EE250 pKa = 4.78GSPEE254 pKa = 3.96HH255 pKa = 7.17KK256 pKa = 9.93PIKK259 pKa = 9.49NVKK262 pKa = 7.71ITYY265 pKa = 8.87DD266 pKa = 2.83GHH268 pKa = 4.93MQIEE272 pKa = 4.38ASVSISAGGNTVVNDD287 pKa = 3.88AVLTNIWTGGTVTVVAGDD305 pKa = 3.85YY306 pKa = 9.75FQLDD310 pKa = 3.4AVIQINAICDD320 pKa = 3.75EE321 pKa = 4.62DD322 pKa = 4.32TLSPTITNWSVDD334 pKa = 3.28DD335 pKa = 3.85ATNQFFNIATFKK347 pKa = 9.57HH348 pKa = 5.72TNPLDD353 pKa = 4.24NNDD356 pKa = 3.71GAHH359 pKa = 6.81LSGSPDD365 pKa = 3.41FPSYY369 pKa = 10.39WIVTEE374 pKa = 3.82ITGDD378 pKa = 3.78LLIVNWLDD386 pKa = 3.3QYY388 pKa = 11.4IFQSDD393 pKa = 3.61NDD395 pKa = 3.73VGIVSSSGVTTEE407 pKa = 5.51IIAGDD412 pKa = 3.76NLAVNTISIMEE423 pKa = 4.57LAYY426 pKa = 10.25AYY428 pKa = 9.97DD429 pKa = 3.99IIVIGGSWYY438 pKa = 10.07DD439 pKa = 3.42ANIIQQLNVLYY450 pKa = 10.92DD451 pKa = 3.5NDD453 pKa = 5.37LIGAVNGFQTTGEE466 pKa = 4.41GSVSTSGNLLWNEE479 pKa = 3.57ALIYY483 pKa = 10.44SVSGDD488 pKa = 3.8YY489 pKa = 11.54GDD491 pKa = 4.5MPAHH495 pKa = 5.2YY496 pKa = 10.11QEE498 pKa = 5.47LADD501 pKa = 4.26NLAAGKK507 pKa = 9.6EE508 pKa = 4.1VLPSGVLSDD517 pKa = 3.9PALAGNEE524 pKa = 4.06ALRR527 pKa = 11.84VLYY530 pKa = 10.43IEE532 pKa = 5.38GDD534 pKa = 4.41LINLNYY540 pKa = 9.51IRR542 pKa = 11.84QTSIVGDD549 pKa = 3.66SDD551 pKa = 4.24QIALAMNEE559 pKa = 3.95FTPRR563 pKa = 11.84ADD565 pKa = 3.94AEE567 pKa = 4.27WTITTGGNALLNHH580 pKa = 6.78AAIVDD585 pKa = 4.42LDD587 pKa = 3.88SLGNIHH593 pKa = 6.87VGGEE597 pKa = 4.14QYY599 pKa = 10.71SQAMLVQAEE608 pKa = 4.85LISDD612 pKa = 3.82KK613 pKa = 11.24PEE615 pKa = 3.84FWAQDD620 pKa = 3.15PDD622 pKa = 3.84ALVNEE627 pKa = 4.75AVAFLDD633 pKa = 5.13DD634 pKa = 5.52SIDD637 pKa = 4.76DD638 pKa = 4.57GPDD641 pKa = 4.28DD642 pKa = 3.88IPAYY646 pKa = 8.74YY647 pKa = 10.28QPDD650 pKa = 3.56NEE652 pKa = 4.49APGGDD657 pKa = 3.95GLQAMLGG664 pKa = 3.68
Molecular weight: 71.6 kDa
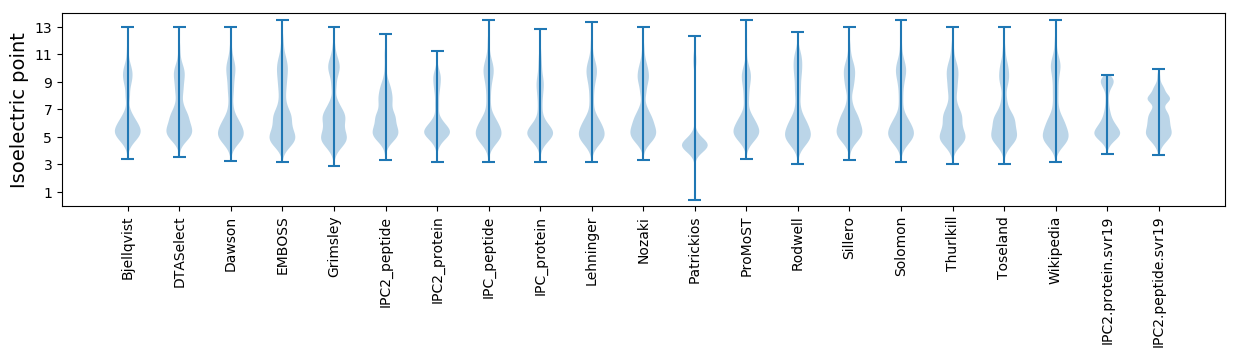
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A562ERA1|A0A562ERA1_9RHIZ O-phosphoserine phosphohydrolase OS=Aminobacter sp. J15 OX=935260 GN=L611_000700000540 PE=3 SV=1
MM1 pKa = 6.74XHH3 pKa = 7.36LSLRR7 pKa = 11.84PKK9 pKa = 10.65SRR11 pKa = 11.84LQLTNPHH18 pKa = 6.33HH19 pKa = 6.72RR20 pKa = 11.84LKK22 pKa = 10.93NPPNNRR28 pKa = 11.84LRR30 pKa = 11.84NNPRR34 pKa = 11.84LKK36 pKa = 10.25RR37 pKa = 11.84QPHH40 pKa = 6.19HH41 pKa = 6.27PMKK44 pKa = 10.16PRR46 pKa = 11.84HH47 pKa = 5.48QRR49 pKa = 11.84LLLKK53 pKa = 10.09QHH55 pKa = 6.4LRR57 pKa = 11.84MPLLSLRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84PRR68 pKa = 11.84KK69 pKa = 9.03PRR71 pKa = 11.84KK72 pKa = 8.86QRR74 pKa = 11.84LRR76 pKa = 11.84QLPRR80 pKa = 11.84PMPLHH85 pKa = 6.36LRR87 pKa = 11.84QEE89 pKa = 4.08KK90 pKa = 9.76RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84HH95 pKa = 5.49LPRR98 pKa = 11.84KK99 pKa = 7.24PRR101 pKa = 11.84RR102 pKa = 11.84VARR105 pKa = 11.84LLLRR109 pKa = 11.84NNPLLQQGSRR119 pKa = 11.84KK120 pKa = 9.41ARR122 pKa = 11.84RR123 pKa = 11.84PLLHH127 pKa = 6.89LHH129 pKa = 6.36LQHH132 pKa = 6.95PPLPRR137 pKa = 11.84RR138 pKa = 11.84ALLLRR143 pKa = 11.84QKK145 pKa = 11.26ASLQLRR151 pKa = 11.84KK152 pKa = 10.11APANRR157 pKa = 11.84LATRR161 pKa = 11.84LRR163 pKa = 11.84CSTALRR169 pKa = 11.84KK170 pKa = 9.61LLLPHH175 pKa = 6.14QRR177 pKa = 11.84VASQLRR183 pKa = 11.84MPPNSQHH190 pKa = 7.28RR191 pKa = 11.84LRR193 pKa = 11.84QLLPDD198 pKa = 3.21RR199 pKa = 11.84RR200 pKa = 11.84LPRR203 pKa = 11.84TSRR206 pKa = 11.84HH207 pKa = 5.4SSLSSRR213 pKa = 11.84SSSARR218 pKa = 11.84CAKK221 pKa = 10.39SKK223 pKa = 10.82VSASTISRR231 pKa = 11.84AAASAGRR238 pKa = 11.84APNRR242 pKa = 11.84SASLATARR250 pKa = 4.51
MM1 pKa = 6.74XHH3 pKa = 7.36LSLRR7 pKa = 11.84PKK9 pKa = 10.65SRR11 pKa = 11.84LQLTNPHH18 pKa = 6.33HH19 pKa = 6.72RR20 pKa = 11.84LKK22 pKa = 10.93NPPNNRR28 pKa = 11.84LRR30 pKa = 11.84NNPRR34 pKa = 11.84LKK36 pKa = 10.25RR37 pKa = 11.84QPHH40 pKa = 6.19HH41 pKa = 6.27PMKK44 pKa = 10.16PRR46 pKa = 11.84HH47 pKa = 5.48QRR49 pKa = 11.84LLLKK53 pKa = 10.09QHH55 pKa = 6.4LRR57 pKa = 11.84MPLLSLRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84PRR68 pKa = 11.84KK69 pKa = 9.03PRR71 pKa = 11.84KK72 pKa = 8.86QRR74 pKa = 11.84LRR76 pKa = 11.84QLPRR80 pKa = 11.84PMPLHH85 pKa = 6.36LRR87 pKa = 11.84QEE89 pKa = 4.08KK90 pKa = 9.76RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84HH95 pKa = 5.49LPRR98 pKa = 11.84KK99 pKa = 7.24PRR101 pKa = 11.84RR102 pKa = 11.84VARR105 pKa = 11.84LLLRR109 pKa = 11.84NNPLLQQGSRR119 pKa = 11.84KK120 pKa = 9.41ARR122 pKa = 11.84RR123 pKa = 11.84PLLHH127 pKa = 6.89LHH129 pKa = 6.36LQHH132 pKa = 6.95PPLPRR137 pKa = 11.84RR138 pKa = 11.84ALLLRR143 pKa = 11.84QKK145 pKa = 11.26ASLQLRR151 pKa = 11.84KK152 pKa = 10.11APANRR157 pKa = 11.84LATRR161 pKa = 11.84LRR163 pKa = 11.84CSTALRR169 pKa = 11.84KK170 pKa = 9.61LLLPHH175 pKa = 6.14QRR177 pKa = 11.84VASQLRR183 pKa = 11.84MPPNSQHH190 pKa = 7.28RR191 pKa = 11.84LRR193 pKa = 11.84QLLPDD198 pKa = 3.21RR199 pKa = 11.84RR200 pKa = 11.84LPRR203 pKa = 11.84TSRR206 pKa = 11.84HH207 pKa = 5.4SSLSSRR213 pKa = 11.84SSSARR218 pKa = 11.84CAKK221 pKa = 10.39SKK223 pKa = 10.82VSASTISRR231 pKa = 11.84AAASAGRR238 pKa = 11.84APNRR242 pKa = 11.84SASLATARR250 pKa = 4.51
Molecular weight: 29.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1237581 |
20 |
2817 |
309.5 |
33.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.917 ± 0.046 | 0.783 ± 0.012 |
5.481 ± 0.035 | 6.389 ± 0.036 |
3.88 ± 0.028 | 8.352 ± 0.037 |
1.991 ± 0.017 | 5.601 ± 0.029 |
3.515 ± 0.029 | 9.842 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.59 ± 0.018 | 2.766 ± 0.022 |
5.01 ± 0.029 | 3.08 ± 0.023 |
7.034 ± 0.041 | 5.45 ± 0.027 |
5.159 ± 0.028 | 7.547 ± 0.035 |
1.289 ± 0.016 | 2.307 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
