
Aphis glycines virus 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.48
Get precalculated fractions of proteins
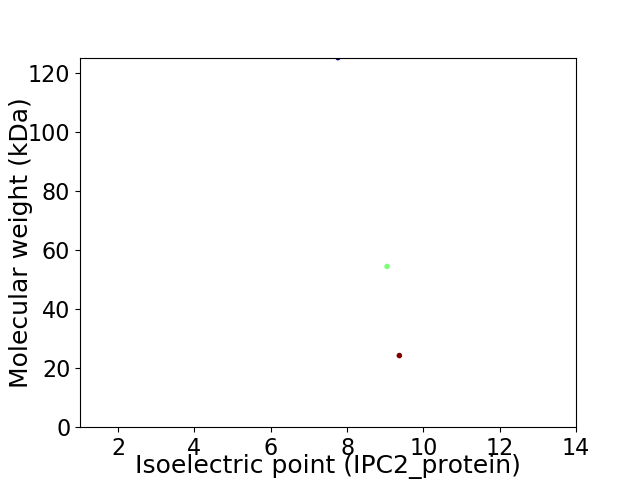
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
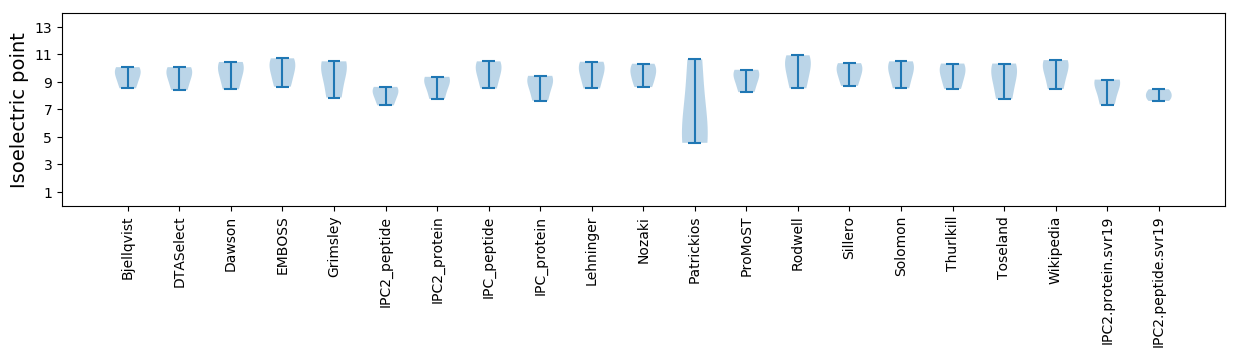
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S1MNC2|A0A0S1MNC2_9VIRU Putative readthrough protein OS=Aphis glycines virus 2 OX=1897732 PE=3 SV=1
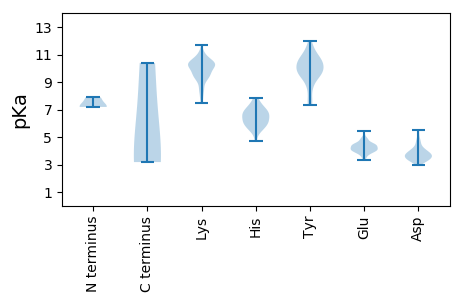
MM1 pKa = 7.88DD2 pKa = 5.5CSNPVNVNDD11 pKa = 5.4RR12 pKa = 11.84ITVGDD17 pKa = 3.87ARR19 pKa = 11.84IAAAGYY25 pKa = 9.48IRR27 pKa = 11.84QDD29 pKa = 3.33LASLKK34 pKa = 10.18NAAAKK39 pKa = 10.36VPRR42 pKa = 11.84NYY44 pKa = 10.69LPTSTPRR51 pKa = 11.84PPDD54 pKa = 3.32VVRR57 pKa = 11.84ADD59 pKa = 3.8LLRR62 pKa = 11.84VKK64 pKa = 9.69TSRR67 pKa = 11.84RR68 pKa = 11.84TVPNDD73 pKa = 3.33PEE75 pKa = 4.24GLARR79 pKa = 11.84LNDD82 pKa = 3.6LSEE85 pKa = 4.2FLTLEE90 pKa = 4.26AEE92 pKa = 4.62SIPICGYY99 pKa = 10.08VGHH102 pKa = 6.59QGNVHH107 pKa = 6.18PSGVKK112 pKa = 8.7MKK114 pKa = 10.28QGRR117 pKa = 11.84SLATRR122 pKa = 11.84TFGGQTRR129 pKa = 11.84DD130 pKa = 4.11DD131 pKa = 3.87IVLEE135 pKa = 3.8AALRR139 pKa = 11.84HH140 pKa = 5.49FPYY143 pKa = 9.76MDD145 pKa = 2.94VAAAYY150 pKa = 9.98SSAVYY155 pKa = 9.76TSGTPSGFLEE165 pKa = 4.08RR166 pKa = 11.84MRR168 pKa = 11.84LNMTRR173 pKa = 11.84PAVSSLRR180 pKa = 11.84TANVRR185 pKa = 11.84NIPQLAALMEE195 pKa = 4.49RR196 pKa = 11.84LLPIRR201 pKa = 11.84KK202 pKa = 9.45APVPDD207 pKa = 3.47WNVDD211 pKa = 3.95FDD213 pKa = 4.15VLLDD217 pKa = 3.76DD218 pKa = 5.15VEE220 pKa = 4.37ITKK223 pKa = 10.29EE224 pKa = 3.71AGAGYY229 pKa = 8.61PYY231 pKa = 10.39CRR233 pKa = 11.84SKK235 pKa = 10.48GQAMRR240 pKa = 11.84DD241 pKa = 3.71CLEE244 pKa = 3.94VVLPNIVEE252 pKa = 4.23AVTTGTWEE260 pKa = 3.86QLMVDD265 pKa = 3.73QPEE268 pKa = 4.25LFLVEE273 pKa = 4.39VKK275 pKa = 10.67NKK277 pKa = 9.84QDD279 pKa = 3.19TYY281 pKa = 11.49DD282 pKa = 3.53VEE284 pKa = 4.85KK285 pKa = 11.01LMTKK289 pKa = 8.11TRR291 pKa = 11.84PYY293 pKa = 9.86MNYY296 pKa = 9.46PMHH299 pKa = 6.6FTLFWSFMAQHH310 pKa = 6.08YY311 pKa = 10.0NEE313 pKa = 4.77LMYY316 pKa = 10.93KK317 pKa = 10.3VGDD320 pKa = 3.98GPATSNACGWSMAAGGGKK338 pKa = 9.85RR339 pKa = 11.84KK340 pKa = 9.49IEE342 pKa = 4.23YY343 pKa = 9.9INKK346 pKa = 7.61QLKK349 pKa = 10.18RR350 pKa = 11.84EE351 pKa = 4.02GDD353 pKa = 3.77WFVEE357 pKa = 4.78AYY359 pKa = 10.47SDD361 pKa = 5.09DD362 pKa = 3.45VVIYY366 pKa = 8.27YY367 pKa = 9.26KK368 pKa = 10.3QHH370 pKa = 5.96GEE372 pKa = 4.15VYY374 pKa = 10.13RR375 pKa = 11.84ISPDD379 pKa = 3.43FVQMDD384 pKa = 3.55SSIDD388 pKa = 3.5ADD390 pKa = 3.36IVRR393 pKa = 11.84ARR395 pKa = 11.84LYY397 pKa = 10.55NIKK400 pKa = 10.42KK401 pKa = 9.92LFKK404 pKa = 9.45EE405 pKa = 3.82QHH407 pKa = 6.17GDD409 pKa = 3.85NVWMEE414 pKa = 4.03SMIDD418 pKa = 3.37LWEE421 pKa = 4.72DD422 pKa = 3.22MALDD426 pKa = 3.79PMLIVQGTKK435 pKa = 7.92IWRR438 pKa = 11.84KK439 pKa = 7.7KK440 pKa = 9.75AKK442 pKa = 9.83HH443 pKa = 5.47GMCSGVPDD451 pKa = 3.52TTGINTYY458 pKa = 10.58KK459 pKa = 10.67SVLAYY464 pKa = 10.49HH465 pKa = 6.87KK466 pKa = 9.45MLEE469 pKa = 4.27SHH471 pKa = 7.63DD472 pKa = 4.04PRR474 pKa = 11.84TLTEE478 pKa = 3.73EE479 pKa = 4.5VARR482 pKa = 11.84KK483 pKa = 7.73WMRR486 pKa = 11.84VHH488 pKa = 7.08AGLEE492 pKa = 4.18IKK494 pKa = 10.46EE495 pKa = 4.48GTWTVEE501 pKa = 4.06PANFMAPPDD510 pKa = 4.06EE511 pKa = 4.54PMCPQTFLGVQWMNTLEE528 pKa = 4.36GTVVPYY534 pKa = 10.99LPTEE538 pKa = 3.34KK539 pKa = 9.47WLRR542 pKa = 11.84MLVVPKK548 pKa = 10.33DD549 pKa = 3.69KK550 pKa = 11.31EE551 pKa = 3.85MDD553 pKa = 3.87SYY555 pKa = 11.96SLAAARR561 pKa = 11.84RR562 pKa = 11.84RR563 pKa = 11.84FDD565 pKa = 3.19RR566 pKa = 11.84MRR568 pKa = 11.84GYY570 pKa = 10.81LITGAAFDD578 pKa = 3.69QRR580 pKa = 11.84IVNVINDD587 pKa = 4.1VLNGITPEE595 pKa = 3.81AVIMQVQAGNGKK607 pKa = 10.16GEE609 pKa = 4.02APQEE613 pKa = 4.06GLLLPDD619 pKa = 3.5YY620 pKa = 10.28VYY622 pKa = 10.32PASDD626 pKa = 3.62VVPTRR631 pKa = 11.84QDD633 pKa = 2.95VEE635 pKa = 4.42DD636 pKa = 4.08IYY638 pKa = 11.19RR639 pKa = 11.84AGAVRR644 pKa = 11.84TPWQEE649 pKa = 3.52LFPGVKK655 pKa = 9.77EE656 pKa = 4.45LIAGHH661 pKa = 6.37MDD663 pKa = 3.88LNQRR667 pKa = 11.84KK668 pKa = 8.47FGKK671 pKa = 9.78IKK673 pKa = 10.19PYY675 pKa = 9.46RR676 pKa = 11.84AKK678 pKa = 10.73LAGAGEE684 pKa = 4.48GKK686 pKa = 10.81YY687 pKa = 10.52SDD689 pKa = 4.74FMVLTPPEE697 pKa = 4.35EE698 pKa = 4.34FASPAQNPGISGAPYY713 pKa = 10.51KK714 pKa = 10.28KK715 pKa = 10.42GKK717 pKa = 7.45HH718 pKa = 4.7TKK720 pKa = 9.68PGNKK724 pKa = 9.4CNVVLPDD731 pKa = 3.62GEE733 pKa = 4.46RR734 pKa = 11.84GKK736 pKa = 9.18TMPTVEE742 pKa = 4.91DD743 pKa = 4.47VIDD746 pKa = 3.66HH747 pKa = 7.18ALRR750 pKa = 11.84EE751 pKa = 4.4SAMPLSLLAQKK762 pKa = 10.27LGKK765 pKa = 9.69EE766 pKa = 3.88PHH768 pKa = 5.8EE769 pKa = 4.35TTKK772 pKa = 10.42FVYY775 pKa = 9.97NAGYY779 pKa = 9.68DD780 pKa = 3.37ARR782 pKa = 11.84EE783 pKa = 3.95PLLGVKK789 pKa = 9.78SEE791 pKa = 4.23NLGPALHH798 pKa = 6.67SLFPVKK804 pKa = 10.02EE805 pKa = 3.96RR806 pKa = 11.84LRR808 pKa = 11.84VRR810 pKa = 11.84EE811 pKa = 4.17TKK813 pKa = 10.44MMDD816 pKa = 3.43DD817 pKa = 3.84GKK819 pKa = 8.21TTVAITKK826 pKa = 9.19PDD828 pKa = 3.35TRR830 pKa = 11.84VTPLRR835 pKa = 11.84GTVVHH840 pKa = 7.37DD841 pKa = 4.04EE842 pKa = 4.48FNGEE846 pKa = 3.67ARR848 pKa = 11.84SPPRR852 pKa = 11.84EE853 pKa = 4.1VKK855 pKa = 10.44VIHH858 pKa = 6.63HH859 pKa = 6.65PLVTSSALVGDD870 pKa = 3.98PVSYY874 pKa = 9.74MVHH877 pKa = 7.28LLTKK881 pKa = 9.47ATGEE885 pKa = 3.92NGEE888 pKa = 4.24WSTTTVPTTIEE899 pKa = 3.83AGTAVVFRR907 pKa = 11.84AEE909 pKa = 3.91YY910 pKa = 9.54SHH912 pKa = 7.87AGFTTHH918 pKa = 6.94AVANSIKK925 pKa = 9.99KK926 pKa = 10.15AKK928 pKa = 10.17AFASRR933 pKa = 11.84VMIDD937 pKa = 3.35KK938 pKa = 10.62LIRR941 pKa = 11.84DD942 pKa = 4.4HH943 pKa = 6.51GVPMPSAYY951 pKa = 9.53KK952 pKa = 10.18PPTPSEE958 pKa = 4.23VIEE961 pKa = 4.12VSARR965 pKa = 11.84ARR967 pKa = 11.84AEE969 pKa = 3.73SLQAHH974 pKa = 6.44NNPSWKK980 pKa = 8.41TAKK983 pKa = 10.4NNQAKK988 pKa = 10.27LDD990 pKa = 3.86YY991 pKa = 9.79AQQKK995 pKa = 8.9SAQIDD1000 pKa = 3.49EE1001 pKa = 5.15RR1002 pKa = 11.84IATRR1006 pKa = 11.84LGPDD1010 pKa = 3.08KK1011 pKa = 10.85PLDD1014 pKa = 3.68EE1015 pKa = 4.89LLVDD1019 pKa = 4.27RR1020 pKa = 11.84FEE1022 pKa = 4.98KK1023 pKa = 10.79LATALTDD1030 pKa = 3.76RR1031 pKa = 11.84LNDD1034 pKa = 4.66AIKK1037 pKa = 10.38QSLGISLEE1045 pKa = 4.03VEE1047 pKa = 3.97ALDD1050 pKa = 4.51SRR1052 pKa = 11.84KK1053 pKa = 9.85KK1054 pKa = 10.91SKK1056 pKa = 9.51TKK1058 pKa = 10.73NKK1060 pKa = 8.9STSDD1064 pKa = 3.2KK1065 pKa = 10.5KK1066 pKa = 11.0AKK1068 pKa = 8.45KK1069 pKa = 8.46TNHH1072 pKa = 5.88KK1073 pKa = 9.07TNSDD1077 pKa = 3.46YY1078 pKa = 10.84DD1079 pKa = 3.74TTNQEE1084 pKa = 3.43EE1085 pKa = 4.24SQYY1088 pKa = 11.15YY1089 pKa = 9.03EE1090 pKa = 4.63DD1091 pKa = 4.24YY1092 pKa = 11.8GEE1094 pKa = 4.55GTSNRR1099 pKa = 11.84YY1100 pKa = 7.65GAPYY1104 pKa = 10.19DD1105 pKa = 3.92EE1106 pKa = 5.42EE1107 pKa = 4.42EE1108 pKa = 4.79GPSKK1112 pKa = 10.76KK1113 pKa = 10.45GMRR1116 pKa = 11.84SRR1118 pKa = 11.84RR1119 pKa = 11.84RR1120 pKa = 11.84RR1121 pKa = 3.32
MM1 pKa = 7.88DD2 pKa = 5.5CSNPVNVNDD11 pKa = 5.4RR12 pKa = 11.84ITVGDD17 pKa = 3.87ARR19 pKa = 11.84IAAAGYY25 pKa = 9.48IRR27 pKa = 11.84QDD29 pKa = 3.33LASLKK34 pKa = 10.18NAAAKK39 pKa = 10.36VPRR42 pKa = 11.84NYY44 pKa = 10.69LPTSTPRR51 pKa = 11.84PPDD54 pKa = 3.32VVRR57 pKa = 11.84ADD59 pKa = 3.8LLRR62 pKa = 11.84VKK64 pKa = 9.69TSRR67 pKa = 11.84RR68 pKa = 11.84TVPNDD73 pKa = 3.33PEE75 pKa = 4.24GLARR79 pKa = 11.84LNDD82 pKa = 3.6LSEE85 pKa = 4.2FLTLEE90 pKa = 4.26AEE92 pKa = 4.62SIPICGYY99 pKa = 10.08VGHH102 pKa = 6.59QGNVHH107 pKa = 6.18PSGVKK112 pKa = 8.7MKK114 pKa = 10.28QGRR117 pKa = 11.84SLATRR122 pKa = 11.84TFGGQTRR129 pKa = 11.84DD130 pKa = 4.11DD131 pKa = 3.87IVLEE135 pKa = 3.8AALRR139 pKa = 11.84HH140 pKa = 5.49FPYY143 pKa = 9.76MDD145 pKa = 2.94VAAAYY150 pKa = 9.98SSAVYY155 pKa = 9.76TSGTPSGFLEE165 pKa = 4.08RR166 pKa = 11.84MRR168 pKa = 11.84LNMTRR173 pKa = 11.84PAVSSLRR180 pKa = 11.84TANVRR185 pKa = 11.84NIPQLAALMEE195 pKa = 4.49RR196 pKa = 11.84LLPIRR201 pKa = 11.84KK202 pKa = 9.45APVPDD207 pKa = 3.47WNVDD211 pKa = 3.95FDD213 pKa = 4.15VLLDD217 pKa = 3.76DD218 pKa = 5.15VEE220 pKa = 4.37ITKK223 pKa = 10.29EE224 pKa = 3.71AGAGYY229 pKa = 8.61PYY231 pKa = 10.39CRR233 pKa = 11.84SKK235 pKa = 10.48GQAMRR240 pKa = 11.84DD241 pKa = 3.71CLEE244 pKa = 3.94VVLPNIVEE252 pKa = 4.23AVTTGTWEE260 pKa = 3.86QLMVDD265 pKa = 3.73QPEE268 pKa = 4.25LFLVEE273 pKa = 4.39VKK275 pKa = 10.67NKK277 pKa = 9.84QDD279 pKa = 3.19TYY281 pKa = 11.49DD282 pKa = 3.53VEE284 pKa = 4.85KK285 pKa = 11.01LMTKK289 pKa = 8.11TRR291 pKa = 11.84PYY293 pKa = 9.86MNYY296 pKa = 9.46PMHH299 pKa = 6.6FTLFWSFMAQHH310 pKa = 6.08YY311 pKa = 10.0NEE313 pKa = 4.77LMYY316 pKa = 10.93KK317 pKa = 10.3VGDD320 pKa = 3.98GPATSNACGWSMAAGGGKK338 pKa = 9.85RR339 pKa = 11.84KK340 pKa = 9.49IEE342 pKa = 4.23YY343 pKa = 9.9INKK346 pKa = 7.61QLKK349 pKa = 10.18RR350 pKa = 11.84EE351 pKa = 4.02GDD353 pKa = 3.77WFVEE357 pKa = 4.78AYY359 pKa = 10.47SDD361 pKa = 5.09DD362 pKa = 3.45VVIYY366 pKa = 8.27YY367 pKa = 9.26KK368 pKa = 10.3QHH370 pKa = 5.96GEE372 pKa = 4.15VYY374 pKa = 10.13RR375 pKa = 11.84ISPDD379 pKa = 3.43FVQMDD384 pKa = 3.55SSIDD388 pKa = 3.5ADD390 pKa = 3.36IVRR393 pKa = 11.84ARR395 pKa = 11.84LYY397 pKa = 10.55NIKK400 pKa = 10.42KK401 pKa = 9.92LFKK404 pKa = 9.45EE405 pKa = 3.82QHH407 pKa = 6.17GDD409 pKa = 3.85NVWMEE414 pKa = 4.03SMIDD418 pKa = 3.37LWEE421 pKa = 4.72DD422 pKa = 3.22MALDD426 pKa = 3.79PMLIVQGTKK435 pKa = 7.92IWRR438 pKa = 11.84KK439 pKa = 7.7KK440 pKa = 9.75AKK442 pKa = 9.83HH443 pKa = 5.47GMCSGVPDD451 pKa = 3.52TTGINTYY458 pKa = 10.58KK459 pKa = 10.67SVLAYY464 pKa = 10.49HH465 pKa = 6.87KK466 pKa = 9.45MLEE469 pKa = 4.27SHH471 pKa = 7.63DD472 pKa = 4.04PRR474 pKa = 11.84TLTEE478 pKa = 3.73EE479 pKa = 4.5VARR482 pKa = 11.84KK483 pKa = 7.73WMRR486 pKa = 11.84VHH488 pKa = 7.08AGLEE492 pKa = 4.18IKK494 pKa = 10.46EE495 pKa = 4.48GTWTVEE501 pKa = 4.06PANFMAPPDD510 pKa = 4.06EE511 pKa = 4.54PMCPQTFLGVQWMNTLEE528 pKa = 4.36GTVVPYY534 pKa = 10.99LPTEE538 pKa = 3.34KK539 pKa = 9.47WLRR542 pKa = 11.84MLVVPKK548 pKa = 10.33DD549 pKa = 3.69KK550 pKa = 11.31EE551 pKa = 3.85MDD553 pKa = 3.87SYY555 pKa = 11.96SLAAARR561 pKa = 11.84RR562 pKa = 11.84RR563 pKa = 11.84FDD565 pKa = 3.19RR566 pKa = 11.84MRR568 pKa = 11.84GYY570 pKa = 10.81LITGAAFDD578 pKa = 3.69QRR580 pKa = 11.84IVNVINDD587 pKa = 4.1VLNGITPEE595 pKa = 3.81AVIMQVQAGNGKK607 pKa = 10.16GEE609 pKa = 4.02APQEE613 pKa = 4.06GLLLPDD619 pKa = 3.5YY620 pKa = 10.28VYY622 pKa = 10.32PASDD626 pKa = 3.62VVPTRR631 pKa = 11.84QDD633 pKa = 2.95VEE635 pKa = 4.42DD636 pKa = 4.08IYY638 pKa = 11.19RR639 pKa = 11.84AGAVRR644 pKa = 11.84TPWQEE649 pKa = 3.52LFPGVKK655 pKa = 9.77EE656 pKa = 4.45LIAGHH661 pKa = 6.37MDD663 pKa = 3.88LNQRR667 pKa = 11.84KK668 pKa = 8.47FGKK671 pKa = 9.78IKK673 pKa = 10.19PYY675 pKa = 9.46RR676 pKa = 11.84AKK678 pKa = 10.73LAGAGEE684 pKa = 4.48GKK686 pKa = 10.81YY687 pKa = 10.52SDD689 pKa = 4.74FMVLTPPEE697 pKa = 4.35EE698 pKa = 4.34FASPAQNPGISGAPYY713 pKa = 10.51KK714 pKa = 10.28KK715 pKa = 10.42GKK717 pKa = 7.45HH718 pKa = 4.7TKK720 pKa = 9.68PGNKK724 pKa = 9.4CNVVLPDD731 pKa = 3.62GEE733 pKa = 4.46RR734 pKa = 11.84GKK736 pKa = 9.18TMPTVEE742 pKa = 4.91DD743 pKa = 4.47VIDD746 pKa = 3.66HH747 pKa = 7.18ALRR750 pKa = 11.84EE751 pKa = 4.4SAMPLSLLAQKK762 pKa = 10.27LGKK765 pKa = 9.69EE766 pKa = 3.88PHH768 pKa = 5.8EE769 pKa = 4.35TTKK772 pKa = 10.42FVYY775 pKa = 9.97NAGYY779 pKa = 9.68DD780 pKa = 3.37ARR782 pKa = 11.84EE783 pKa = 3.95PLLGVKK789 pKa = 9.78SEE791 pKa = 4.23NLGPALHH798 pKa = 6.67SLFPVKK804 pKa = 10.02EE805 pKa = 3.96RR806 pKa = 11.84LRR808 pKa = 11.84VRR810 pKa = 11.84EE811 pKa = 4.17TKK813 pKa = 10.44MMDD816 pKa = 3.43DD817 pKa = 3.84GKK819 pKa = 8.21TTVAITKK826 pKa = 9.19PDD828 pKa = 3.35TRR830 pKa = 11.84VTPLRR835 pKa = 11.84GTVVHH840 pKa = 7.37DD841 pKa = 4.04EE842 pKa = 4.48FNGEE846 pKa = 3.67ARR848 pKa = 11.84SPPRR852 pKa = 11.84EE853 pKa = 4.1VKK855 pKa = 10.44VIHH858 pKa = 6.63HH859 pKa = 6.65PLVTSSALVGDD870 pKa = 3.98PVSYY874 pKa = 9.74MVHH877 pKa = 7.28LLTKK881 pKa = 9.47ATGEE885 pKa = 3.92NGEE888 pKa = 4.24WSTTTVPTTIEE899 pKa = 3.83AGTAVVFRR907 pKa = 11.84AEE909 pKa = 3.91YY910 pKa = 9.54SHH912 pKa = 7.87AGFTTHH918 pKa = 6.94AVANSIKK925 pKa = 9.99KK926 pKa = 10.15AKK928 pKa = 10.17AFASRR933 pKa = 11.84VMIDD937 pKa = 3.35KK938 pKa = 10.62LIRR941 pKa = 11.84DD942 pKa = 4.4HH943 pKa = 6.51GVPMPSAYY951 pKa = 9.53KK952 pKa = 10.18PPTPSEE958 pKa = 4.23VIEE961 pKa = 4.12VSARR965 pKa = 11.84ARR967 pKa = 11.84AEE969 pKa = 3.73SLQAHH974 pKa = 6.44NNPSWKK980 pKa = 8.41TAKK983 pKa = 10.4NNQAKK988 pKa = 10.27LDD990 pKa = 3.86YY991 pKa = 9.79AQQKK995 pKa = 8.9SAQIDD1000 pKa = 3.49EE1001 pKa = 5.15RR1002 pKa = 11.84IATRR1006 pKa = 11.84LGPDD1010 pKa = 3.08KK1011 pKa = 10.85PLDD1014 pKa = 3.68EE1015 pKa = 4.89LLVDD1019 pKa = 4.27RR1020 pKa = 11.84FEE1022 pKa = 4.98KK1023 pKa = 10.79LATALTDD1030 pKa = 3.76RR1031 pKa = 11.84LNDD1034 pKa = 4.66AIKK1037 pKa = 10.38QSLGISLEE1045 pKa = 4.03VEE1047 pKa = 3.97ALDD1050 pKa = 4.51SRR1052 pKa = 11.84KK1053 pKa = 9.85KK1054 pKa = 10.91SKK1056 pKa = 9.51TKK1058 pKa = 10.73NKK1060 pKa = 8.9STSDD1064 pKa = 3.2KK1065 pKa = 10.5KK1066 pKa = 11.0AKK1068 pKa = 8.45KK1069 pKa = 8.46TNHH1072 pKa = 5.88KK1073 pKa = 9.07TNSDD1077 pKa = 3.46YY1078 pKa = 10.84DD1079 pKa = 3.74TTNQEE1084 pKa = 3.43EE1085 pKa = 4.24SQYY1088 pKa = 11.15YY1089 pKa = 9.03EE1090 pKa = 4.63DD1091 pKa = 4.24YY1092 pKa = 11.8GEE1094 pKa = 4.55GTSNRR1099 pKa = 11.84YY1100 pKa = 7.65GAPYY1104 pKa = 10.19DD1105 pKa = 3.92EE1106 pKa = 5.42EE1107 pKa = 4.42EE1108 pKa = 4.79GPSKK1112 pKa = 10.76KK1113 pKa = 10.45GMRR1116 pKa = 11.84SRR1118 pKa = 11.84RR1119 pKa = 11.84RR1120 pKa = 11.84RR1121 pKa = 3.32
Molecular weight: 125.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S1MNB6|A0A0S1MNB6_9VIRU Replicase OS=Aphis glycines virus 2 OX=1897732 PE=4 SV=1
MM1 pKa = 7.19TRR3 pKa = 11.84RR4 pKa = 11.84TKK6 pKa = 10.51KK7 pKa = 10.08RR8 pKa = 11.84ANTTKK13 pKa = 10.14TMEE16 pKa = 4.41KK17 pKa = 10.55ALVTAMEE24 pKa = 4.33RR25 pKa = 11.84LTTKK29 pKa = 10.32RR30 pKa = 11.84RR31 pKa = 11.84VPRR34 pKa = 11.84KK35 pKa = 9.41KK36 pKa = 9.19GCARR40 pKa = 11.84VAEE43 pKa = 4.52GEE45 pKa = 4.14MVLRR49 pKa = 11.84RR50 pKa = 11.84EE51 pKa = 4.02EE52 pKa = 3.99MLVDD56 pKa = 3.57VTLSANKK63 pKa = 9.04TDD65 pKa = 3.66STGSVVLALANFPWLKK81 pKa = 9.38TVAGSFEE88 pKa = 4.1RR89 pKa = 11.84YY90 pKa = 7.3KK91 pKa = 10.22WKK93 pKa = 10.28RR94 pKa = 11.84LNIHH98 pKa = 5.54WRR100 pKa = 11.84AAGGFNKK107 pKa = 10.35GGLIAVGMDD116 pKa = 3.27WSNQLSSAYY125 pKa = 8.88TRR127 pKa = 11.84QTLTSASEE135 pKa = 4.08RR136 pKa = 11.84QKK138 pKa = 11.18VLSLTPHH145 pKa = 6.0MSLPISSTSINKK157 pKa = 8.45TLGLPIKK164 pKa = 9.62MLNSRR169 pKa = 11.84NWYY172 pKa = 9.54DD173 pKa = 3.23AAKK176 pKa = 9.48TDD178 pKa = 4.31DD179 pKa = 4.05EE180 pKa = 4.88GAVGAIRR187 pKa = 11.84YY188 pKa = 7.57SAKK191 pKa = 10.15CDD193 pKa = 3.26SDD195 pKa = 3.7TVEE198 pKa = 4.39RR199 pKa = 11.84FIGEE203 pKa = 3.48IWVDD207 pKa = 3.62YY208 pKa = 10.72EE209 pKa = 4.51VVLQGTRR216 pKa = 11.84AA217 pKa = 3.18
MM1 pKa = 7.19TRR3 pKa = 11.84RR4 pKa = 11.84TKK6 pKa = 10.51KK7 pKa = 10.08RR8 pKa = 11.84ANTTKK13 pKa = 10.14TMEE16 pKa = 4.41KK17 pKa = 10.55ALVTAMEE24 pKa = 4.33RR25 pKa = 11.84LTTKK29 pKa = 10.32RR30 pKa = 11.84RR31 pKa = 11.84VPRR34 pKa = 11.84KK35 pKa = 9.41KK36 pKa = 9.19GCARR40 pKa = 11.84VAEE43 pKa = 4.52GEE45 pKa = 4.14MVLRR49 pKa = 11.84RR50 pKa = 11.84EE51 pKa = 4.02EE52 pKa = 3.99MLVDD56 pKa = 3.57VTLSANKK63 pKa = 9.04TDD65 pKa = 3.66STGSVVLALANFPWLKK81 pKa = 9.38TVAGSFEE88 pKa = 4.1RR89 pKa = 11.84YY90 pKa = 7.3KK91 pKa = 10.22WKK93 pKa = 10.28RR94 pKa = 11.84LNIHH98 pKa = 5.54WRR100 pKa = 11.84AAGGFNKK107 pKa = 10.35GGLIAVGMDD116 pKa = 3.27WSNQLSSAYY125 pKa = 8.88TRR127 pKa = 11.84QTLTSASEE135 pKa = 4.08RR136 pKa = 11.84QKK138 pKa = 11.18VLSLTPHH145 pKa = 6.0MSLPISSTSINKK157 pKa = 8.45TLGLPIKK164 pKa = 9.62MLNSRR169 pKa = 11.84NWYY172 pKa = 9.54DD173 pKa = 3.23AAKK176 pKa = 9.48TDD178 pKa = 4.31DD179 pKa = 4.05EE180 pKa = 4.88GAVGAIRR187 pKa = 11.84YY188 pKa = 7.57SAKK191 pKa = 10.15CDD193 pKa = 3.26SDD195 pKa = 3.7TVEE198 pKa = 4.39RR199 pKa = 11.84FIGEE203 pKa = 3.48IWVDD207 pKa = 3.62YY208 pKa = 10.72EE209 pKa = 4.51VVLQGTRR216 pKa = 11.84AA217 pKa = 3.18
Molecular weight: 24.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1823 |
217 |
1121 |
607.7 |
68.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.283 ± 0.337 | 0.768 ± 0.056 |
5.595 ± 0.634 | 5.979 ± 0.504 |
2.743 ± 0.447 | 6.253 ± 0.273 |
1.865 ± 0.366 | 3.73 ± 0.102 |
6.802 ± 0.398 | 8.338 ± 0.635 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.236 ± 0.291 | 4.114 ± 0.011 |
5.705 ± 0.967 | 2.907 ± 0.253 |
6.967 ± 0.648 | 6.308 ± 0.826 |
7.625 ± 0.986 | 7.57 ± 0.18 |
1.591 ± 0.32 | 3.566 ± 0.308 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
