
Flavobacteria bacterium (strain BBFL7)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; unclassified Flavobacteriia
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
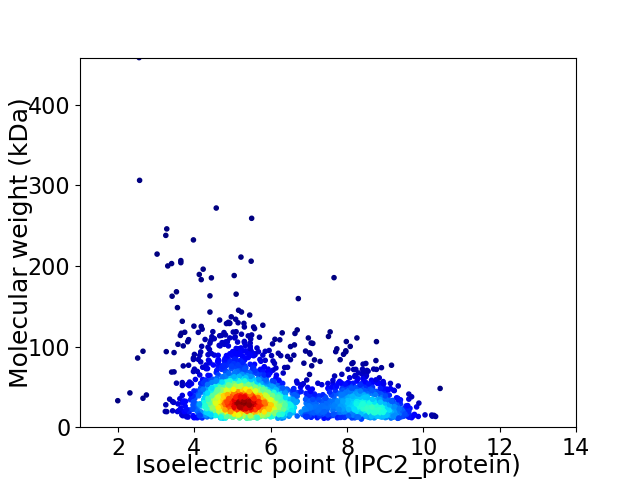
Virtual 2D-PAGE plot for 2592 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
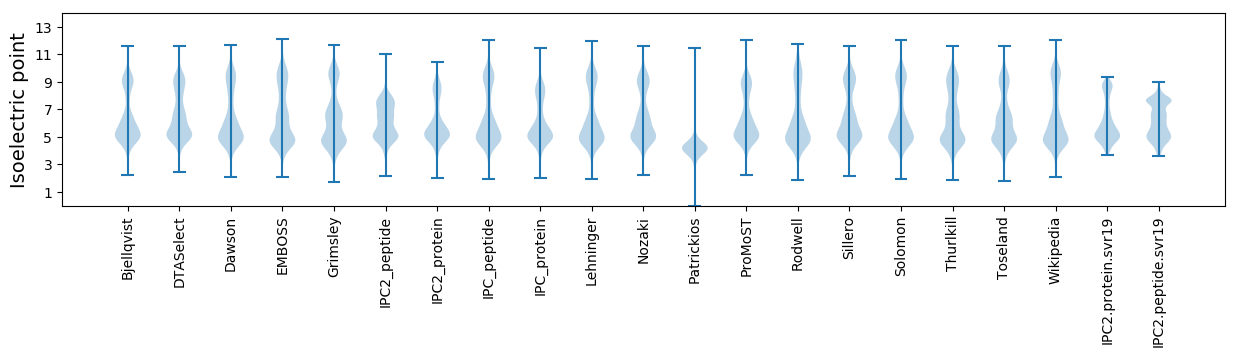
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q26I84|Q26I84_FLABB YfcH-like protein NAD dependent epimerase/dehydratase family OS=Flavobacteria bacterium (strain BBFL7) OX=156586 GN=BBFL7_01164 PE=3 SV=1
MM1 pKa = 7.77KK2 pKa = 10.45KK3 pKa = 9.86PLLLISFLFVGFLSIAQTANDD24 pKa = 4.03FVFDD28 pKa = 4.64SILANMWPFSISSSTGVTIDD48 pKa = 3.4WGDD51 pKa = 3.39GTTTNVPMGNNQTVPAHH68 pKa = 6.13SFSSTTRR75 pKa = 11.84TVIISGSIDD84 pKa = 3.51SIVFNLSSSSNNNPRR99 pKa = 11.84ITQWGSSQWKK109 pKa = 5.4TTEE112 pKa = 3.53WMFFDD117 pKa = 5.55CLYY120 pKa = 11.18LDD122 pKa = 4.54INASDD127 pKa = 3.94TPDD130 pKa = 3.91LSLCNSMAHH139 pKa = 6.02MFHH142 pKa = 7.29NAGQFNDD149 pKa = 5.7DD150 pKa = 3.34INNWDD155 pKa = 3.52VSNITDD161 pKa = 3.8MSDD164 pKa = 2.83LFYY167 pKa = 11.13GVYY170 pKa = 10.46SFNQALNNWDD180 pKa = 3.58VSSVTDD186 pKa = 3.29MSRR189 pKa = 11.84MFLYY193 pKa = 10.63ASNFNQPLSNWDD205 pKa = 3.42TANVINMGSMFQAASSFNSNINNWDD230 pKa = 3.24VSNVSNISHH239 pKa = 6.78MFYY242 pKa = 10.57NANSFNQDD250 pKa = 2.04ISNWDD255 pKa = 3.4VSNVNAIDD263 pKa = 3.62YY264 pKa = 9.92LFYY267 pKa = 10.77RR268 pKa = 11.84ATSFNQNISNWDD280 pKa = 3.23VSNITNMDD288 pKa = 3.22RR289 pKa = 11.84MFTIASSFNQNISSWDD305 pKa = 3.54VSNVTSMIEE314 pKa = 3.76MFEE317 pKa = 4.23YY318 pKa = 9.35ATSFNQPIDD327 pKa = 3.29NWDD330 pKa = 3.55VSNVTIMQDD339 pKa = 3.28MFVHH343 pKa = 5.96ATSFNQNISSWDD355 pKa = 3.5VSNVTDD361 pKa = 4.26MDD363 pKa = 4.17DD364 pKa = 3.16MFYY367 pKa = 11.03NATSFNQPLNNWDD380 pKa = 3.39FNQYY384 pKa = 10.26LNPRR388 pKa = 11.84RR389 pKa = 11.84FVAFSGLDD397 pKa = 3.24TNNYY401 pKa = 9.64DD402 pKa = 3.66KK403 pKa = 11.29LLQRR407 pKa = 11.84FVNLGMQYY415 pKa = 8.76KK416 pKa = 8.73TLNSFGLTYY425 pKa = 10.54CDD427 pKa = 3.07SFSRR431 pKa = 11.84NILLQNGWSINGDD444 pKa = 3.94SLDD447 pKa = 4.52LGCSALDD454 pKa = 3.58NQLQGSITYY463 pKa = 10.6DD464 pKa = 3.51FDD466 pKa = 6.0NNGCDD471 pKa = 3.69PNDD474 pKa = 4.53LIFSNVGIEE483 pKa = 3.99ISNGTTAFTLYY494 pKa = 10.54TDD496 pKa = 3.18INGDD500 pKa = 3.49YY501 pKa = 10.61SIPLSDD507 pKa = 3.33NTYY510 pKa = 8.94TVTPIIDD517 pKa = 3.66TNLFTTTPASATFTAANQNITTQDD541 pKa = 3.1FCLTATTPTDD551 pKa = 3.59DD552 pKa = 5.16LEE554 pKa = 4.18ITIIPLEE561 pKa = 4.05EE562 pKa = 4.1ARR564 pKa = 11.84PGFDD568 pKa = 2.83TNYY571 pKa = 10.82KK572 pKa = 10.08LVYY575 pKa = 9.26KK576 pKa = 10.87NKK578 pKa = 10.41GNTQLTGNIAFTYY591 pKa = 10.67DD592 pKa = 3.65DD593 pKa = 4.7DD594 pKa = 5.73FMDD597 pKa = 4.96FVSANPVVSSSSVGNLNWSFSNLDD621 pKa = 3.35PFEE624 pKa = 4.05TRR626 pKa = 11.84EE627 pKa = 3.91IDD629 pKa = 3.26FTLNMNTPTDD639 pKa = 3.78SNFPLNSNDD648 pKa = 3.54IISFTASITPSANDD662 pKa = 3.42ATPIDD667 pKa = 3.8NVFNLDD673 pKa = 3.48QTVVNSYY680 pKa = 11.02DD681 pKa = 3.92PNDD684 pKa = 3.69KK685 pKa = 9.89TCLQGEE691 pKa = 4.53TIEE694 pKa = 4.13PSMVGEE700 pKa = 4.32FVHH703 pKa = 6.22YY704 pKa = 10.22RR705 pKa = 11.84IRR707 pKa = 11.84FEE709 pKa = 4.37NEE711 pKa = 3.02GTASAINVRR720 pKa = 11.84IVDD723 pKa = 4.15YY724 pKa = 10.86IDD726 pKa = 3.34TSVFDD731 pKa = 4.39ISTLTPVSSSHH742 pKa = 7.63DD743 pKa = 3.73YY744 pKa = 10.51LAQITDD750 pKa = 3.6GNKK753 pKa = 10.37LEE755 pKa = 5.5FIFDD759 pKa = 4.27NINLPNTAPASQGYY773 pKa = 9.58ILFKK777 pKa = 10.6IKK779 pKa = 9.7TVDD782 pKa = 3.45TLVLGDD788 pKa = 3.98TFSNQAGIYY797 pKa = 9.9FDD799 pKa = 4.81FNFPIITNLEE809 pKa = 4.2TTTVAVTLSNNDD821 pKa = 2.76ITSLTMSLFPNPANDD836 pKa = 3.51TVTITSNVLFDD847 pKa = 3.31SCTIYY852 pKa = 10.94DD853 pKa = 4.12LRR855 pKa = 11.84GMLISSKK862 pKa = 10.57KK863 pKa = 9.77LQQPDD868 pKa = 3.44DD869 pKa = 3.68QYY871 pKa = 12.03KK872 pKa = 10.71FNIDD876 pKa = 3.34SLASGMYY883 pKa = 9.31MIQINNSQGSITQRR897 pKa = 11.84LIVEE901 pKa = 4.57
MM1 pKa = 7.77KK2 pKa = 10.45KK3 pKa = 9.86PLLLISFLFVGFLSIAQTANDD24 pKa = 4.03FVFDD28 pKa = 4.64SILANMWPFSISSSTGVTIDD48 pKa = 3.4WGDD51 pKa = 3.39GTTTNVPMGNNQTVPAHH68 pKa = 6.13SFSSTTRR75 pKa = 11.84TVIISGSIDD84 pKa = 3.51SIVFNLSSSSNNNPRR99 pKa = 11.84ITQWGSSQWKK109 pKa = 5.4TTEE112 pKa = 3.53WMFFDD117 pKa = 5.55CLYY120 pKa = 11.18LDD122 pKa = 4.54INASDD127 pKa = 3.94TPDD130 pKa = 3.91LSLCNSMAHH139 pKa = 6.02MFHH142 pKa = 7.29NAGQFNDD149 pKa = 5.7DD150 pKa = 3.34INNWDD155 pKa = 3.52VSNITDD161 pKa = 3.8MSDD164 pKa = 2.83LFYY167 pKa = 11.13GVYY170 pKa = 10.46SFNQALNNWDD180 pKa = 3.58VSSVTDD186 pKa = 3.29MSRR189 pKa = 11.84MFLYY193 pKa = 10.63ASNFNQPLSNWDD205 pKa = 3.42TANVINMGSMFQAASSFNSNINNWDD230 pKa = 3.24VSNVSNISHH239 pKa = 6.78MFYY242 pKa = 10.57NANSFNQDD250 pKa = 2.04ISNWDD255 pKa = 3.4VSNVNAIDD263 pKa = 3.62YY264 pKa = 9.92LFYY267 pKa = 10.77RR268 pKa = 11.84ATSFNQNISNWDD280 pKa = 3.23VSNITNMDD288 pKa = 3.22RR289 pKa = 11.84MFTIASSFNQNISSWDD305 pKa = 3.54VSNVTSMIEE314 pKa = 3.76MFEE317 pKa = 4.23YY318 pKa = 9.35ATSFNQPIDD327 pKa = 3.29NWDD330 pKa = 3.55VSNVTIMQDD339 pKa = 3.28MFVHH343 pKa = 5.96ATSFNQNISSWDD355 pKa = 3.5VSNVTDD361 pKa = 4.26MDD363 pKa = 4.17DD364 pKa = 3.16MFYY367 pKa = 11.03NATSFNQPLNNWDD380 pKa = 3.39FNQYY384 pKa = 10.26LNPRR388 pKa = 11.84RR389 pKa = 11.84FVAFSGLDD397 pKa = 3.24TNNYY401 pKa = 9.64DD402 pKa = 3.66KK403 pKa = 11.29LLQRR407 pKa = 11.84FVNLGMQYY415 pKa = 8.76KK416 pKa = 8.73TLNSFGLTYY425 pKa = 10.54CDD427 pKa = 3.07SFSRR431 pKa = 11.84NILLQNGWSINGDD444 pKa = 3.94SLDD447 pKa = 4.52LGCSALDD454 pKa = 3.58NQLQGSITYY463 pKa = 10.6DD464 pKa = 3.51FDD466 pKa = 6.0NNGCDD471 pKa = 3.69PNDD474 pKa = 4.53LIFSNVGIEE483 pKa = 3.99ISNGTTAFTLYY494 pKa = 10.54TDD496 pKa = 3.18INGDD500 pKa = 3.49YY501 pKa = 10.61SIPLSDD507 pKa = 3.33NTYY510 pKa = 8.94TVTPIIDD517 pKa = 3.66TNLFTTTPASATFTAANQNITTQDD541 pKa = 3.1FCLTATTPTDD551 pKa = 3.59DD552 pKa = 5.16LEE554 pKa = 4.18ITIIPLEE561 pKa = 4.05EE562 pKa = 4.1ARR564 pKa = 11.84PGFDD568 pKa = 2.83TNYY571 pKa = 10.82KK572 pKa = 10.08LVYY575 pKa = 9.26KK576 pKa = 10.87NKK578 pKa = 10.41GNTQLTGNIAFTYY591 pKa = 10.67DD592 pKa = 3.65DD593 pKa = 4.7DD594 pKa = 5.73FMDD597 pKa = 4.96FVSANPVVSSSSVGNLNWSFSNLDD621 pKa = 3.35PFEE624 pKa = 4.05TRR626 pKa = 11.84EE627 pKa = 3.91IDD629 pKa = 3.26FTLNMNTPTDD639 pKa = 3.78SNFPLNSNDD648 pKa = 3.54IISFTASITPSANDD662 pKa = 3.42ATPIDD667 pKa = 3.8NVFNLDD673 pKa = 3.48QTVVNSYY680 pKa = 11.02DD681 pKa = 3.92PNDD684 pKa = 3.69KK685 pKa = 9.89TCLQGEE691 pKa = 4.53TIEE694 pKa = 4.13PSMVGEE700 pKa = 4.32FVHH703 pKa = 6.22YY704 pKa = 10.22RR705 pKa = 11.84IRR707 pKa = 11.84FEE709 pKa = 4.37NEE711 pKa = 3.02GTASAINVRR720 pKa = 11.84IVDD723 pKa = 4.15YY724 pKa = 10.86IDD726 pKa = 3.34TSVFDD731 pKa = 4.39ISTLTPVSSSHH742 pKa = 7.63DD743 pKa = 3.73YY744 pKa = 10.51LAQITDD750 pKa = 3.6GNKK753 pKa = 10.37LEE755 pKa = 5.5FIFDD759 pKa = 4.27NINLPNTAPASQGYY773 pKa = 9.58ILFKK777 pKa = 10.6IKK779 pKa = 9.7TVDD782 pKa = 3.45TLVLGDD788 pKa = 3.98TFSNQAGIYY797 pKa = 9.9FDD799 pKa = 4.81FNFPIITNLEE809 pKa = 4.2TTTVAVTLSNNDD821 pKa = 2.76ITSLTMSLFPNPANDD836 pKa = 3.51TVTITSNVLFDD847 pKa = 3.31SCTIYY852 pKa = 10.94DD853 pKa = 4.12LRR855 pKa = 11.84GMLISSKK862 pKa = 10.57KK863 pKa = 9.77LQQPDD868 pKa = 3.44DD869 pKa = 3.68QYY871 pKa = 12.03KK872 pKa = 10.71FNIDD876 pKa = 3.34SLASGMYY883 pKa = 9.31MIQINNSQGSITQRR897 pKa = 11.84LIVEE901 pKa = 4.57
Molecular weight: 100.82 kDa
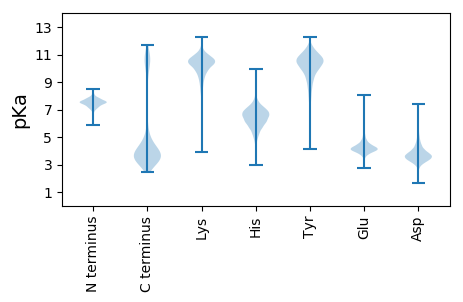
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q26D32|Q26D32_FLABB 30S ribosomal protein S3 OS=Flavobacteria bacterium (strain BBFL7) OX=156586 GN=rpsC PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.72KK28 pKa = 11.01PEE30 pKa = 4.39KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.34RR39 pKa = 11.84SGGRR43 pKa = 11.84NASGRR48 pKa = 11.84MTMRR52 pKa = 11.84YY53 pKa = 9.61KK54 pKa = 10.94GGGHH58 pKa = 5.71KK59 pKa = 9.74RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.82FKK68 pKa = 10.86RR69 pKa = 11.84DD70 pKa = 3.53KK71 pKa = 10.96QGVPATIASIEE82 pKa = 3.99YY83 pKa = 10.19DD84 pKa = 3.48PNRR87 pKa = 11.84TAFIALVNYY96 pKa = 9.88QDD98 pKa = 3.39GEE100 pKa = 4.04KK101 pKa = 10.2RR102 pKa = 11.84YY103 pKa = 10.72VIAQNGLQVGQNIVSGDD120 pKa = 3.53SVAPEE125 pKa = 3.84VGNAMKK131 pKa = 10.58LSSIPLGSIISCIEE145 pKa = 3.55LHH147 pKa = 6.68PGQGAVIARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.33YY171 pKa = 9.08ATVKK175 pKa = 9.73MPSGEE180 pKa = 4.13TRR182 pKa = 11.84LVLQEE187 pKa = 4.05CLATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.63AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.3GYY255 pKa = 7.46RR256 pKa = 11.84TRR258 pKa = 11.84DD259 pKa = 3.23KK260 pKa = 11.38NKK262 pKa = 10.48ASTQYY267 pKa = 10.43ILEE270 pKa = 4.16RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.21KK274 pKa = 9.94
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.72KK28 pKa = 11.01PEE30 pKa = 4.39KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.34RR39 pKa = 11.84SGGRR43 pKa = 11.84NASGRR48 pKa = 11.84MTMRR52 pKa = 11.84YY53 pKa = 9.61KK54 pKa = 10.94GGGHH58 pKa = 5.71KK59 pKa = 9.74RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.82FKK68 pKa = 10.86RR69 pKa = 11.84DD70 pKa = 3.53KK71 pKa = 10.96QGVPATIASIEE82 pKa = 3.99YY83 pKa = 10.19DD84 pKa = 3.48PNRR87 pKa = 11.84TAFIALVNYY96 pKa = 9.88QDD98 pKa = 3.39GEE100 pKa = 4.04KK101 pKa = 10.2RR102 pKa = 11.84YY103 pKa = 10.72VIAQNGLQVGQNIVSGDD120 pKa = 3.53SVAPEE125 pKa = 3.84VGNAMKK131 pKa = 10.58LSSIPLGSIISCIEE145 pKa = 3.55LHH147 pKa = 6.68PGQGAVIARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.33YY171 pKa = 9.08ATVKK175 pKa = 9.73MPSGEE180 pKa = 4.13TRR182 pKa = 11.84LVLQEE187 pKa = 4.05CLATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.63AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.3GYY255 pKa = 7.46RR256 pKa = 11.84TRR258 pKa = 11.84DD259 pKa = 3.23KK260 pKa = 11.38NKK262 pKa = 10.48ASTQYY267 pKa = 10.43ILEE270 pKa = 4.16RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.21KK274 pKa = 9.94
Molecular weight: 29.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
928669 |
88 |
4500 |
358.3 |
40.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.754 ± 0.043 | 0.746 ± 0.021 |
6.328 ± 0.068 | 6.235 ± 0.047 |
4.843 ± 0.031 | 6.495 ± 0.049 |
1.879 ± 0.029 | 7.784 ± 0.048 |
6.561 ± 0.072 | 9.092 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.025 | 5.988 ± 0.05 |
3.429 ± 0.033 | 3.733 ± 0.028 |
3.623 ± 0.035 | 6.583 ± 0.048 |
6.123 ± 0.085 | 6.306 ± 0.036 |
1.041 ± 0.016 | 4.085 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
