
Zymomonas mobilis subsp. mobilis (strain ATCC 31821 / ZM4 / CP4)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Zymomonadaceae; Zymomonas; Zymomonas mobilis; Zymomonas mobilis subsp. mobilis
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
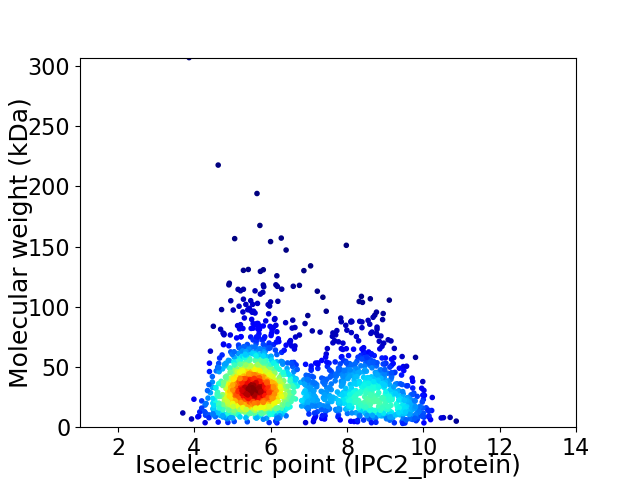
Virtual 2D-PAGE plot for 1779 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5NR00|Q5NR00_ZYMMO ABC transporter related protein OS=Zymomonas mobilis subsp. mobilis (strain ATCC 31821 / ZM4 / CP4) OX=264203 GN=ZMO0230 PE=4 SV=1
MM1 pKa = 7.64ASSLTITKK9 pKa = 10.16KK10 pKa = 10.43RR11 pKa = 11.84GNHH14 pKa = 5.62KK15 pKa = 10.64AFSTEE20 pKa = 3.81EE21 pKa = 3.74NHH23 pKa = 7.32ISLQHH28 pKa = 5.83PSVVQVNAYY37 pKa = 9.26RR38 pKa = 11.84SQVRR42 pKa = 11.84SLSRR46 pKa = 11.84QGDD49 pKa = 3.8TLLVQLNDD57 pKa = 2.9GRR59 pKa = 11.84QYY61 pKa = 11.04QFDD64 pKa = 3.99NFFVDD69 pKa = 4.53DD70 pKa = 4.74DD71 pKa = 4.05KK72 pKa = 11.59HH73 pKa = 6.51LHH75 pKa = 6.14SDD77 pKa = 3.8LVLRR81 pKa = 11.84DD82 pKa = 3.79EE83 pKa = 4.69DD84 pKa = 3.93QKK86 pKa = 10.63EE87 pKa = 4.37AWWHH91 pKa = 6.04AEE93 pKa = 4.05MPDD96 pKa = 3.4ASHH99 pKa = 7.42PNLAGADD106 pKa = 3.06IGYY109 pKa = 8.84SQVEE113 pKa = 4.65TIKK116 pKa = 10.81PLLIYY121 pKa = 10.0HH122 pKa = 7.25DD123 pKa = 4.4NGLLVPLLAGLLGAGAAGGAAAALTGGHH151 pKa = 5.31SHH153 pKa = 7.52KK154 pKa = 11.06DD155 pKa = 3.36NNNNSDD161 pKa = 3.64LSIPSVDD168 pKa = 4.51SVTTDD173 pKa = 3.23PTTGEE178 pKa = 4.07VTVNGTAQPGQTVNVTFPDD197 pKa = 3.6GTVAQTTADD206 pKa = 3.52SKK208 pKa = 11.6GDD210 pKa = 3.48YY211 pKa = 9.63HH212 pKa = 6.41ATSTTPQATGQIAVTSSDD230 pKa = 3.03SSSNVSNPVTHH241 pKa = 8.01DD242 pKa = 3.47YY243 pKa = 11.49TDD245 pKa = 3.17ITPAIVITSDD255 pKa = 3.39AGNTVQTGPVTYY267 pKa = 10.05TFTLSEE273 pKa = 4.0ASKK276 pKa = 11.15DD277 pKa = 3.43FTTASVTVTGGQKK290 pKa = 10.83GEE292 pKa = 4.14LVQDD296 pKa = 3.71SSNPLVYY303 pKa = 10.68HH304 pKa = 5.42MTVTPDD310 pKa = 2.97AGKK313 pKa = 10.62GGTITVVVPAGSVHH327 pKa = 6.91DD328 pKa = 4.66LSGSTTTADD337 pKa = 3.27AVATPVAYY345 pKa = 7.73DD346 pKa = 3.43TTPAGNPVVTVTSDD360 pKa = 3.38SDD362 pKa = 3.69GQTATGPVTYY372 pKa = 10.0SFKK375 pKa = 10.78LSEE378 pKa = 4.29PVSDD382 pKa = 4.12LTADD386 pKa = 3.5SVTVTGGSKK395 pKa = 10.46GALVQDD401 pKa = 3.73STDD404 pKa = 3.31PLVYY408 pKa = 10.65HH409 pKa = 6.15MVVTPDD415 pKa = 3.16GSKK418 pKa = 10.9GGTVTATIASGSLHH432 pKa = 7.18DD433 pKa = 4.76LAGNPTLSDD442 pKa = 3.23ATDD445 pKa = 3.38TVAYY449 pKa = 7.6DD450 pKa = 3.48TTPAGNPVVTVTSDD464 pKa = 3.38SDD466 pKa = 3.69GQTATGPVTYY476 pKa = 10.0SFKK479 pKa = 10.78LSEE482 pKa = 4.29PVSDD486 pKa = 4.12LTADD490 pKa = 3.5SVTVTGGSKK499 pKa = 10.46GALVQDD505 pKa = 3.73STDD508 pKa = 3.31PLVYY512 pKa = 10.65HH513 pKa = 6.15MVVTPDD519 pKa = 3.16GSKK522 pKa = 10.9GGTVTATIASGSLHH536 pKa = 7.18DD537 pKa = 4.76LAGNPTLSDD546 pKa = 3.23ATDD549 pKa = 3.38TVAYY553 pKa = 7.6DD554 pKa = 3.48TTPAGNPVVTVTSDD568 pKa = 3.38SDD570 pKa = 3.69GQTATGPVTYY580 pKa = 10.0SFKK583 pKa = 10.78LSEE586 pKa = 4.29PVSDD590 pKa = 4.12LTADD594 pKa = 3.5SVTVTGGSKK603 pKa = 10.47GALVQDD609 pKa = 3.69TTDD612 pKa = 3.41PLVYY616 pKa = 10.55HH617 pKa = 6.09MVVTPDD623 pKa = 3.16GSKK626 pKa = 10.9GGTVTATIASGSLHH640 pKa = 7.18DD641 pKa = 4.76LAGNPTLSDD650 pKa = 3.23ATDD653 pKa = 3.38TVAYY657 pKa = 7.6DD658 pKa = 3.48TTPAGNPVVTVTSDD672 pKa = 3.38SDD674 pKa = 3.69GQTATGPVTYY684 pKa = 10.0SFKK687 pKa = 10.78LSEE690 pKa = 4.35PVSDD694 pKa = 3.9LTTDD698 pKa = 3.21SVTVTGGSKK707 pKa = 10.46GALVQDD713 pKa = 3.73STDD716 pKa = 3.31PLVYY720 pKa = 10.65HH721 pKa = 6.15MVVTPDD727 pKa = 3.16GSKK730 pKa = 10.9GGTVTATIASGSLHH744 pKa = 7.18DD745 pKa = 4.76LAGNPTLSDD754 pKa = 3.23ATDD757 pKa = 3.38TVAYY761 pKa = 7.6DD762 pKa = 3.48TTPAGNPVVTVTSDD776 pKa = 3.38SDD778 pKa = 3.69GQTATGPVTYY788 pKa = 10.0SFKK791 pKa = 10.78LSEE794 pKa = 4.35PVSDD798 pKa = 3.9LTTDD802 pKa = 3.21SVTVTGGSKK811 pKa = 10.47GALVQDD817 pKa = 3.69TTDD820 pKa = 3.41PLVYY824 pKa = 10.55HH825 pKa = 6.09MVVTPDD831 pKa = 3.16GSKK834 pKa = 10.93GGTITATIASGSLHH848 pKa = 7.18DD849 pKa = 4.76LAGNPTLSDD858 pKa = 3.23ATDD861 pKa = 3.38TVAYY865 pKa = 7.6DD866 pKa = 3.48TTPAGNPVVTVTSDD880 pKa = 3.38SDD882 pKa = 3.69GQTATGPVTYY892 pKa = 10.0SFKK895 pKa = 10.78LSEE898 pKa = 4.29PVSDD902 pKa = 4.12LTADD906 pKa = 3.5SVTVTGGSKK915 pKa = 10.47GALVQDD921 pKa = 3.69TTDD924 pKa = 3.41PLVYY928 pKa = 10.52HH929 pKa = 6.01MVVTPEE935 pKa = 3.91TGTGGTVSVTVPKK948 pKa = 10.74GAIHH952 pKa = 7.38DD953 pKa = 4.19LAGNPTLSDD962 pKa = 3.1ATDD965 pKa = 3.12NVAYY969 pKa = 7.83DD970 pKa = 3.69TTPAGNPTLSVTSTHH985 pKa = 7.09DD986 pKa = 3.15GVAATNTPVTFGFIASEE1003 pKa = 3.94PLKK1006 pKa = 10.96GFSADD1011 pKa = 3.65NIEE1014 pKa = 4.33VTGGTKK1020 pKa = 10.61GEE1022 pKa = 4.26LTSSPDD1028 pKa = 3.18HH1029 pKa = 6.59PLMYY1033 pKa = 10.53FLTVTPNPNSSGEE1046 pKa = 3.88ISIKK1050 pKa = 10.45VPAGAVFDD1058 pKa = 3.94MAGNPNLSSVNDD1070 pKa = 3.67TVLYY1074 pKa = 8.89DD1075 pKa = 4.16TIPPTVTISDD1085 pKa = 3.85DD1086 pKa = 4.18SNGKK1090 pKa = 6.32TVNGAVHH1097 pKa = 5.01YY1098 pKa = 7.7TFTASEE1104 pKa = 4.11PLLAFAADD1112 pKa = 4.01NIEE1115 pKa = 4.19VTGGTKK1121 pKa = 10.38GDD1123 pKa = 3.88LVQDD1127 pKa = 3.75SSNPLVYY1134 pKa = 10.76HH1135 pKa = 5.44MTVTPNSASGGSIDD1149 pKa = 3.77VKK1151 pKa = 10.57IPAGSVHH1158 pKa = 7.24DD1159 pKa = 4.14LAGNPTASDD1168 pKa = 3.49VSDD1171 pKa = 3.47TVAYY1175 pKa = 7.69DD1176 pKa = 3.36TTNIDD1181 pKa = 3.73SPVVTITSDD1190 pKa = 3.31SDD1192 pKa = 3.45GKK1194 pKa = 9.87ATAEE1198 pKa = 3.75PVTFKK1203 pKa = 10.39FTLSEE1208 pKa = 4.07PLSDD1212 pKa = 4.74FNADD1216 pKa = 3.18SVTVSGGTKK1225 pKa = 10.41GNLIQDD1231 pKa = 4.07TSNPLVYY1238 pKa = 10.95YY1239 pKa = 7.18MTVTPKK1245 pKa = 9.53VTVSPMTIDD1254 pKa = 3.34TPSALSATVSAGSVHH1269 pKa = 6.24NLAGNPTLSDD1279 pKa = 3.32ATDD1282 pKa = 3.52SVPYY1286 pKa = 10.04DD1287 pKa = 3.32SSVATNPIVTITSDD1301 pKa = 3.17SDD1303 pKa = 3.48GQTVIGPVTYY1313 pKa = 10.61NFKK1316 pKa = 10.77LSQAVNDD1323 pKa = 4.08FTADD1327 pKa = 3.36SVTVTGGSKK1336 pKa = 10.49GQLVQDD1342 pKa = 3.77SSDD1345 pKa = 3.3PLLYY1349 pKa = 10.92HH1350 pKa = 6.21MVVTPDD1356 pKa = 3.15ANQTGNVTATIPSGQLHH1373 pKa = 6.51NIIGLSNLGNSTDD1386 pKa = 4.05SIAFDD1391 pKa = 3.64TAPPTIAITSTGDD1404 pKa = 3.13NDD1406 pKa = 3.76TTTEE1410 pKa = 3.93GTITYY1415 pKa = 7.37TFTLSEE1421 pKa = 4.0PLKK1424 pKa = 11.08NFTADD1429 pKa = 4.14DD1430 pKa = 3.73INVYY1434 pKa = 10.64GGTKK1438 pKa = 10.19GSLIQDD1444 pKa = 3.58SSNPLSYY1451 pKa = 11.22SLVVTPNPNSGGTLVVNIPAGSVQDD1476 pKa = 3.85VNGNPMVVDD1485 pKa = 3.63STHH1488 pKa = 5.67AVGYY1492 pKa = 9.85IIQPNDD1498 pKa = 2.86VRR1500 pKa = 11.84VFSDD1504 pKa = 3.25QDD1506 pKa = 3.16GSTATGDD1513 pKa = 2.74ITYY1516 pKa = 10.63YY1517 pKa = 11.06VVFNNSNPAFSPNDD1531 pKa = 3.34LTIEE1535 pKa = 4.09NGTLKK1540 pKa = 10.73SFEE1543 pKa = 4.29RR1544 pKa = 11.84TSGSADD1550 pKa = 3.34GTNAAYY1556 pKa = 9.58KK1557 pKa = 10.39IIVTPVADD1565 pKa = 3.45NAGINSLSISPDD1577 pKa = 3.05VTNGSGVITGMVVFDD1592 pKa = 3.58TRR1594 pKa = 11.84QLNLQDD1600 pKa = 3.77YY1601 pKa = 8.6TDD1603 pKa = 4.12TGASANDD1610 pKa = 4.48YY1611 pKa = 10.45ISQDD1615 pKa = 2.94NSFTLNLDD1623 pKa = 3.14NAANGTTPSYY1633 pKa = 10.71QISKK1637 pKa = 10.98DD1638 pKa = 3.53NGTTWTDD1645 pKa = 3.25TSATQSNLPDD1655 pKa = 3.07GHH1657 pKa = 6.46YY1658 pKa = 10.52LYY1660 pKa = 10.66RR1661 pKa = 11.84AVRR1664 pKa = 11.84NDD1666 pKa = 3.23VHH1668 pKa = 7.78DD1669 pKa = 3.95MPLYY1673 pKa = 11.09SNVIDD1678 pKa = 3.98LTVDD1682 pKa = 3.12TTKK1685 pKa = 10.85PDD1687 pKa = 4.42LISINPTNGQTVSGIAEE1704 pKa = 4.36AGTTVNLLSNDD1715 pKa = 3.27GQTVLGTVVADD1726 pKa = 4.65SNNHH1730 pKa = 4.35WSISGLNIADD1740 pKa = 3.66GTPIKK1745 pKa = 10.18ATATDD1750 pKa = 3.41TAGNVSSVVTTRR1762 pKa = 11.84VDD1764 pKa = 3.32ATPPAVTVDD1773 pKa = 3.54NSNGTTITGTTEE1785 pKa = 3.14AGAVVKK1791 pKa = 10.81VDD1793 pKa = 3.56TNGDD1797 pKa = 3.21GVADD1801 pKa = 3.73YY1802 pKa = 11.17TEE1804 pKa = 4.49FAGSDD1809 pKa = 3.98GKK1811 pKa = 10.27WSVRR1815 pKa = 11.84PDD1817 pKa = 3.17TPLANGTSVSVTSTDD1832 pKa = 3.2PVGNVSAAATTVVDD1846 pKa = 3.77NVAPVVTLITADD1858 pKa = 3.56NQTLTGTVDD1867 pKa = 3.43DD1868 pKa = 4.32STVTSVKK1875 pKa = 10.4VFDD1878 pKa = 3.54NTGNFVGTGTVTNGSYY1894 pKa = 11.02SLTLADD1900 pKa = 4.11PVAPGTVLTAKK1911 pKa = 9.71ATDD1914 pKa = 3.47PAGNVGSSSSLTIGSVSSDD1933 pKa = 3.05SDD1935 pKa = 3.36NSALATHH1942 pKa = 6.17QAAITAIATDD1952 pKa = 4.11TLPTSAVSTVQNNDD1966 pKa = 2.34FDD1968 pKa = 4.36TRR1970 pKa = 11.84DD1971 pKa = 3.48PTLTVSGTSNLEE1983 pKa = 3.89PAQEE1987 pKa = 3.53AAAASFIAMPLAMSLTSSNASSSTFSDD2014 pKa = 3.52VLQISTDD2021 pKa = 3.43GQNWSTVTPSANGAWTWTDD2040 pKa = 3.37PTAHH2044 pKa = 7.36DD2045 pKa = 4.72SNFTYY2050 pKa = 10.69YY2051 pKa = 10.82LRR2053 pKa = 11.84TLDD2056 pKa = 3.45SSGKK2060 pKa = 9.71VVYY2063 pKa = 7.62TTSKK2067 pKa = 10.94DD2068 pKa = 3.12ITIDD2072 pKa = 4.3LIAPSSLVMPTWNDD2086 pKa = 2.87AGITVTKK2093 pKa = 10.7DD2094 pKa = 2.6ADD2096 pKa = 3.59ATVALINDD2104 pKa = 3.73VNHH2107 pKa = 7.13DD2108 pKa = 3.52GVYY2111 pKa = 9.93EE2112 pKa = 3.99QGVDD2116 pKa = 3.25KK2117 pKa = 11.19VLAVADD2123 pKa = 3.98NSGTTATLSATLTAGQHH2140 pKa = 5.38YY2141 pKa = 10.56NLGLVQYY2148 pKa = 9.79DD2149 pKa = 3.38VAGNYY2154 pKa = 9.57SRR2156 pKa = 11.84LSNTVSYY2163 pKa = 7.61TAPPTSALNTAFQASVLPPQEE2184 pKa = 4.28VRR2186 pKa = 11.84DD2187 pKa = 4.03SAGMAVGFDD2196 pKa = 3.43AYY2198 pKa = 10.92GNLNILQRR2206 pKa = 11.84SSLITQSNSTNYY2218 pKa = 10.58ALSDD2222 pKa = 3.88VQQLPYY2228 pKa = 10.48LVRR2231 pKa = 11.84YY2232 pKa = 6.81TNQSTDD2238 pKa = 2.53SRR2240 pKa = 11.84NTINSSTIVDD2250 pKa = 3.91YY2251 pKa = 11.57NRR2253 pKa = 11.84DD2254 pKa = 3.47GYY2256 pKa = 11.56SDD2258 pKa = 4.04VLASDD2263 pKa = 4.98YY2264 pKa = 10.84IDD2266 pKa = 3.59SATTGVAVWTGSQSGYY2282 pKa = 10.26SLTRR2286 pKa = 11.84ATSSNNSAGGVMAYY2300 pKa = 10.36DD2301 pKa = 3.87KK2302 pKa = 11.49DD2303 pKa = 3.55GDD2305 pKa = 4.27GYY2307 pKa = 11.56VDD2309 pKa = 3.86AVLGNWGPQNGATNGTFIKK2328 pKa = 9.32NTNGVLSQDD2337 pKa = 3.78GTNGTAGLQNFQFEE2351 pKa = 4.67RR2352 pKa = 11.84EE2353 pKa = 4.09VSGVDD2358 pKa = 3.27LKK2360 pKa = 11.55GVGNIDD2366 pKa = 3.53VAAHH2370 pKa = 6.62TISNSASSNQYY2381 pKa = 11.23ALALIDD2387 pKa = 4.12NNGGTLSLGQNIDD2400 pKa = 3.08NVFNSRR2406 pKa = 11.84GIPTGTTGQKK2416 pKa = 10.39ALPSVWGAQSMVWADD2431 pKa = 3.74FANNGHH2437 pKa = 6.4MDD2439 pKa = 4.07LYY2441 pKa = 10.99LSQGGNTPSGSDD2453 pKa = 3.25NTDD2456 pKa = 2.45SRR2458 pKa = 11.84IYY2460 pKa = 9.95MNNNGVLSSTPTIIADD2476 pKa = 3.64DD2477 pKa = 4.01NLAGGAAIATDD2488 pKa = 3.65WNHH2491 pKa = 6.84DD2492 pKa = 3.36GKK2494 pKa = 10.75IDD2496 pKa = 3.85VIEE2499 pKa = 4.0IRR2501 pKa = 11.84TAIEE2505 pKa = 3.94NNVTNIPVTLLTNNGVNSNGTLNPFTVSNMTTIPRR2540 pKa = 11.84ANITGIAATDD2550 pKa = 3.89VNWDD2554 pKa = 3.53GAVDD2558 pKa = 4.32LLYY2561 pKa = 10.31STASGSDD2568 pKa = 3.27TQADD2572 pKa = 3.89KK2573 pKa = 11.3AGKK2576 pKa = 5.95TTEE2579 pKa = 4.04GNIYY2583 pKa = 10.54SIMNTNAVADD2593 pKa = 4.26GTSLHH2598 pKa = 6.93LKK2600 pKa = 10.1ILDD2603 pKa = 3.69QNGVNSYY2610 pKa = 10.53FGNTVQLFDD2619 pKa = 4.32SDD2621 pKa = 4.24GNLVSTQIINPQEE2634 pKa = 4.09GMWNNDD2640 pKa = 2.11SSGIVNFYY2648 pKa = 11.16NLDD2651 pKa = 3.88PNQHH2655 pKa = 5.68YY2656 pKa = 10.62SVALVRR2662 pKa = 11.84ATDD2665 pKa = 3.42GHH2667 pKa = 6.96SNDD2670 pKa = 2.84IGGRR2674 pKa = 11.84DD2675 pKa = 3.13SYY2677 pKa = 12.03GQFFTLSSVEE2687 pKa = 4.32DD2688 pKa = 3.63SSSVSDD2694 pKa = 3.87GGSEE2698 pKa = 3.84TGAASALNTVEE2709 pKa = 4.52NVEE2712 pKa = 4.25KK2713 pKa = 10.43SWNNLTPGAANHH2725 pKa = 6.61AYY2727 pKa = 10.12VLEE2730 pKa = 4.83GDD2732 pKa = 3.8TTASHH2737 pKa = 5.14TTGGIFTGTGYY2748 pKa = 11.12DD2749 pKa = 3.3DD2750 pKa = 3.76TFFAGAGNNSYY2761 pKa = 10.45IGGGGWGYY2769 pKa = 11.23NSAGEE2774 pKa = 4.22YY2775 pKa = 10.22VWMSSGGHH2783 pKa = 5.24NVVDD2787 pKa = 3.81YY2788 pKa = 10.85SGEE2791 pKa = 4.02SSAITVNMGNSVGIGTVSVSKK2812 pKa = 10.02TDD2814 pKa = 3.25GHH2816 pKa = 7.27DD2817 pKa = 3.51SLQSIQGIVGTNYY2830 pKa = 10.62DD2831 pKa = 4.54DD2832 pKa = 5.7SFTSDD2837 pKa = 2.61GKK2839 pKa = 11.23GDD2841 pKa = 3.69YY2842 pKa = 10.89FEE2844 pKa = 6.45GGGGNDD2850 pKa = 3.43TFNLAKK2856 pKa = 10.39GGHH2859 pKa = 5.99DD2860 pKa = 2.84TLMYY2864 pKa = 10.82KK2865 pKa = 10.72LLDD2868 pKa = 3.86NSDD2871 pKa = 3.59NTGGNGSDD2879 pKa = 3.56TVNDD2883 pKa = 3.49FHH2885 pKa = 8.75LGNTVNNPDD2894 pKa = 4.0ADD2896 pKa = 3.96IINIKK2901 pKa = 10.35DD2902 pKa = 3.82LLADD2906 pKa = 3.72YY2907 pKa = 10.79QGTANVFYY2915 pKa = 10.81DD2916 pKa = 3.34VTEE2919 pKa = 4.22NKK2921 pKa = 9.82FVMDD2925 pKa = 4.34KK2926 pKa = 10.97ASSGLDD2932 pKa = 3.12QFVSTSVHH2940 pKa = 6.05NGNTTISVDD2949 pKa = 3.23RR2950 pKa = 11.84DD2951 pKa = 3.5GANGNHH2957 pKa = 5.83QMTALVTLDD2966 pKa = 3.35NTQTDD2971 pKa = 4.3LATLIGNHH2979 pKa = 5.57QLIIAA2984 pKa = 4.8
MM1 pKa = 7.64ASSLTITKK9 pKa = 10.16KK10 pKa = 10.43RR11 pKa = 11.84GNHH14 pKa = 5.62KK15 pKa = 10.64AFSTEE20 pKa = 3.81EE21 pKa = 3.74NHH23 pKa = 7.32ISLQHH28 pKa = 5.83PSVVQVNAYY37 pKa = 9.26RR38 pKa = 11.84SQVRR42 pKa = 11.84SLSRR46 pKa = 11.84QGDD49 pKa = 3.8TLLVQLNDD57 pKa = 2.9GRR59 pKa = 11.84QYY61 pKa = 11.04QFDD64 pKa = 3.99NFFVDD69 pKa = 4.53DD70 pKa = 4.74DD71 pKa = 4.05KK72 pKa = 11.59HH73 pKa = 6.51LHH75 pKa = 6.14SDD77 pKa = 3.8LVLRR81 pKa = 11.84DD82 pKa = 3.79EE83 pKa = 4.69DD84 pKa = 3.93QKK86 pKa = 10.63EE87 pKa = 4.37AWWHH91 pKa = 6.04AEE93 pKa = 4.05MPDD96 pKa = 3.4ASHH99 pKa = 7.42PNLAGADD106 pKa = 3.06IGYY109 pKa = 8.84SQVEE113 pKa = 4.65TIKK116 pKa = 10.81PLLIYY121 pKa = 10.0HH122 pKa = 7.25DD123 pKa = 4.4NGLLVPLLAGLLGAGAAGGAAAALTGGHH151 pKa = 5.31SHH153 pKa = 7.52KK154 pKa = 11.06DD155 pKa = 3.36NNNNSDD161 pKa = 3.64LSIPSVDD168 pKa = 4.51SVTTDD173 pKa = 3.23PTTGEE178 pKa = 4.07VTVNGTAQPGQTVNVTFPDD197 pKa = 3.6GTVAQTTADD206 pKa = 3.52SKK208 pKa = 11.6GDD210 pKa = 3.48YY211 pKa = 9.63HH212 pKa = 6.41ATSTTPQATGQIAVTSSDD230 pKa = 3.03SSSNVSNPVTHH241 pKa = 8.01DD242 pKa = 3.47YY243 pKa = 11.49TDD245 pKa = 3.17ITPAIVITSDD255 pKa = 3.39AGNTVQTGPVTYY267 pKa = 10.05TFTLSEE273 pKa = 4.0ASKK276 pKa = 11.15DD277 pKa = 3.43FTTASVTVTGGQKK290 pKa = 10.83GEE292 pKa = 4.14LVQDD296 pKa = 3.71SSNPLVYY303 pKa = 10.68HH304 pKa = 5.42MTVTPDD310 pKa = 2.97AGKK313 pKa = 10.62GGTITVVVPAGSVHH327 pKa = 6.91DD328 pKa = 4.66LSGSTTTADD337 pKa = 3.27AVATPVAYY345 pKa = 7.73DD346 pKa = 3.43TTPAGNPVVTVTSDD360 pKa = 3.38SDD362 pKa = 3.69GQTATGPVTYY372 pKa = 10.0SFKK375 pKa = 10.78LSEE378 pKa = 4.29PVSDD382 pKa = 4.12LTADD386 pKa = 3.5SVTVTGGSKK395 pKa = 10.46GALVQDD401 pKa = 3.73STDD404 pKa = 3.31PLVYY408 pKa = 10.65HH409 pKa = 6.15MVVTPDD415 pKa = 3.16GSKK418 pKa = 10.9GGTVTATIASGSLHH432 pKa = 7.18DD433 pKa = 4.76LAGNPTLSDD442 pKa = 3.23ATDD445 pKa = 3.38TVAYY449 pKa = 7.6DD450 pKa = 3.48TTPAGNPVVTVTSDD464 pKa = 3.38SDD466 pKa = 3.69GQTATGPVTYY476 pKa = 10.0SFKK479 pKa = 10.78LSEE482 pKa = 4.29PVSDD486 pKa = 4.12LTADD490 pKa = 3.5SVTVTGGSKK499 pKa = 10.46GALVQDD505 pKa = 3.73STDD508 pKa = 3.31PLVYY512 pKa = 10.65HH513 pKa = 6.15MVVTPDD519 pKa = 3.16GSKK522 pKa = 10.9GGTVTATIASGSLHH536 pKa = 7.18DD537 pKa = 4.76LAGNPTLSDD546 pKa = 3.23ATDD549 pKa = 3.38TVAYY553 pKa = 7.6DD554 pKa = 3.48TTPAGNPVVTVTSDD568 pKa = 3.38SDD570 pKa = 3.69GQTATGPVTYY580 pKa = 10.0SFKK583 pKa = 10.78LSEE586 pKa = 4.29PVSDD590 pKa = 4.12LTADD594 pKa = 3.5SVTVTGGSKK603 pKa = 10.47GALVQDD609 pKa = 3.69TTDD612 pKa = 3.41PLVYY616 pKa = 10.55HH617 pKa = 6.09MVVTPDD623 pKa = 3.16GSKK626 pKa = 10.9GGTVTATIASGSLHH640 pKa = 7.18DD641 pKa = 4.76LAGNPTLSDD650 pKa = 3.23ATDD653 pKa = 3.38TVAYY657 pKa = 7.6DD658 pKa = 3.48TTPAGNPVVTVTSDD672 pKa = 3.38SDD674 pKa = 3.69GQTATGPVTYY684 pKa = 10.0SFKK687 pKa = 10.78LSEE690 pKa = 4.35PVSDD694 pKa = 3.9LTTDD698 pKa = 3.21SVTVTGGSKK707 pKa = 10.46GALVQDD713 pKa = 3.73STDD716 pKa = 3.31PLVYY720 pKa = 10.65HH721 pKa = 6.15MVVTPDD727 pKa = 3.16GSKK730 pKa = 10.9GGTVTATIASGSLHH744 pKa = 7.18DD745 pKa = 4.76LAGNPTLSDD754 pKa = 3.23ATDD757 pKa = 3.38TVAYY761 pKa = 7.6DD762 pKa = 3.48TTPAGNPVVTVTSDD776 pKa = 3.38SDD778 pKa = 3.69GQTATGPVTYY788 pKa = 10.0SFKK791 pKa = 10.78LSEE794 pKa = 4.35PVSDD798 pKa = 3.9LTTDD802 pKa = 3.21SVTVTGGSKK811 pKa = 10.47GALVQDD817 pKa = 3.69TTDD820 pKa = 3.41PLVYY824 pKa = 10.55HH825 pKa = 6.09MVVTPDD831 pKa = 3.16GSKK834 pKa = 10.93GGTITATIASGSLHH848 pKa = 7.18DD849 pKa = 4.76LAGNPTLSDD858 pKa = 3.23ATDD861 pKa = 3.38TVAYY865 pKa = 7.6DD866 pKa = 3.48TTPAGNPVVTVTSDD880 pKa = 3.38SDD882 pKa = 3.69GQTATGPVTYY892 pKa = 10.0SFKK895 pKa = 10.78LSEE898 pKa = 4.29PVSDD902 pKa = 4.12LTADD906 pKa = 3.5SVTVTGGSKK915 pKa = 10.47GALVQDD921 pKa = 3.69TTDD924 pKa = 3.41PLVYY928 pKa = 10.52HH929 pKa = 6.01MVVTPEE935 pKa = 3.91TGTGGTVSVTVPKK948 pKa = 10.74GAIHH952 pKa = 7.38DD953 pKa = 4.19LAGNPTLSDD962 pKa = 3.1ATDD965 pKa = 3.12NVAYY969 pKa = 7.83DD970 pKa = 3.69TTPAGNPTLSVTSTHH985 pKa = 7.09DD986 pKa = 3.15GVAATNTPVTFGFIASEE1003 pKa = 3.94PLKK1006 pKa = 10.96GFSADD1011 pKa = 3.65NIEE1014 pKa = 4.33VTGGTKK1020 pKa = 10.61GEE1022 pKa = 4.26LTSSPDD1028 pKa = 3.18HH1029 pKa = 6.59PLMYY1033 pKa = 10.53FLTVTPNPNSSGEE1046 pKa = 3.88ISIKK1050 pKa = 10.45VPAGAVFDD1058 pKa = 3.94MAGNPNLSSVNDD1070 pKa = 3.67TVLYY1074 pKa = 8.89DD1075 pKa = 4.16TIPPTVTISDD1085 pKa = 3.85DD1086 pKa = 4.18SNGKK1090 pKa = 6.32TVNGAVHH1097 pKa = 5.01YY1098 pKa = 7.7TFTASEE1104 pKa = 4.11PLLAFAADD1112 pKa = 4.01NIEE1115 pKa = 4.19VTGGTKK1121 pKa = 10.38GDD1123 pKa = 3.88LVQDD1127 pKa = 3.75SSNPLVYY1134 pKa = 10.76HH1135 pKa = 5.44MTVTPNSASGGSIDD1149 pKa = 3.77VKK1151 pKa = 10.57IPAGSVHH1158 pKa = 7.24DD1159 pKa = 4.14LAGNPTASDD1168 pKa = 3.49VSDD1171 pKa = 3.47TVAYY1175 pKa = 7.69DD1176 pKa = 3.36TTNIDD1181 pKa = 3.73SPVVTITSDD1190 pKa = 3.31SDD1192 pKa = 3.45GKK1194 pKa = 9.87ATAEE1198 pKa = 3.75PVTFKK1203 pKa = 10.39FTLSEE1208 pKa = 4.07PLSDD1212 pKa = 4.74FNADD1216 pKa = 3.18SVTVSGGTKK1225 pKa = 10.41GNLIQDD1231 pKa = 4.07TSNPLVYY1238 pKa = 10.95YY1239 pKa = 7.18MTVTPKK1245 pKa = 9.53VTVSPMTIDD1254 pKa = 3.34TPSALSATVSAGSVHH1269 pKa = 6.24NLAGNPTLSDD1279 pKa = 3.32ATDD1282 pKa = 3.52SVPYY1286 pKa = 10.04DD1287 pKa = 3.32SSVATNPIVTITSDD1301 pKa = 3.17SDD1303 pKa = 3.48GQTVIGPVTYY1313 pKa = 10.61NFKK1316 pKa = 10.77LSQAVNDD1323 pKa = 4.08FTADD1327 pKa = 3.36SVTVTGGSKK1336 pKa = 10.49GQLVQDD1342 pKa = 3.77SSDD1345 pKa = 3.3PLLYY1349 pKa = 10.92HH1350 pKa = 6.21MVVTPDD1356 pKa = 3.15ANQTGNVTATIPSGQLHH1373 pKa = 6.51NIIGLSNLGNSTDD1386 pKa = 4.05SIAFDD1391 pKa = 3.64TAPPTIAITSTGDD1404 pKa = 3.13NDD1406 pKa = 3.76TTTEE1410 pKa = 3.93GTITYY1415 pKa = 7.37TFTLSEE1421 pKa = 4.0PLKK1424 pKa = 11.08NFTADD1429 pKa = 4.14DD1430 pKa = 3.73INVYY1434 pKa = 10.64GGTKK1438 pKa = 10.19GSLIQDD1444 pKa = 3.58SSNPLSYY1451 pKa = 11.22SLVVTPNPNSGGTLVVNIPAGSVQDD1476 pKa = 3.85VNGNPMVVDD1485 pKa = 3.63STHH1488 pKa = 5.67AVGYY1492 pKa = 9.85IIQPNDD1498 pKa = 2.86VRR1500 pKa = 11.84VFSDD1504 pKa = 3.25QDD1506 pKa = 3.16GSTATGDD1513 pKa = 2.74ITYY1516 pKa = 10.63YY1517 pKa = 11.06VVFNNSNPAFSPNDD1531 pKa = 3.34LTIEE1535 pKa = 4.09NGTLKK1540 pKa = 10.73SFEE1543 pKa = 4.29RR1544 pKa = 11.84TSGSADD1550 pKa = 3.34GTNAAYY1556 pKa = 9.58KK1557 pKa = 10.39IIVTPVADD1565 pKa = 3.45NAGINSLSISPDD1577 pKa = 3.05VTNGSGVITGMVVFDD1592 pKa = 3.58TRR1594 pKa = 11.84QLNLQDD1600 pKa = 3.77YY1601 pKa = 8.6TDD1603 pKa = 4.12TGASANDD1610 pKa = 4.48YY1611 pKa = 10.45ISQDD1615 pKa = 2.94NSFTLNLDD1623 pKa = 3.14NAANGTTPSYY1633 pKa = 10.71QISKK1637 pKa = 10.98DD1638 pKa = 3.53NGTTWTDD1645 pKa = 3.25TSATQSNLPDD1655 pKa = 3.07GHH1657 pKa = 6.46YY1658 pKa = 10.52LYY1660 pKa = 10.66RR1661 pKa = 11.84AVRR1664 pKa = 11.84NDD1666 pKa = 3.23VHH1668 pKa = 7.78DD1669 pKa = 3.95MPLYY1673 pKa = 11.09SNVIDD1678 pKa = 3.98LTVDD1682 pKa = 3.12TTKK1685 pKa = 10.85PDD1687 pKa = 4.42LISINPTNGQTVSGIAEE1704 pKa = 4.36AGTTVNLLSNDD1715 pKa = 3.27GQTVLGTVVADD1726 pKa = 4.65SNNHH1730 pKa = 4.35WSISGLNIADD1740 pKa = 3.66GTPIKK1745 pKa = 10.18ATATDD1750 pKa = 3.41TAGNVSSVVTTRR1762 pKa = 11.84VDD1764 pKa = 3.32ATPPAVTVDD1773 pKa = 3.54NSNGTTITGTTEE1785 pKa = 3.14AGAVVKK1791 pKa = 10.81VDD1793 pKa = 3.56TNGDD1797 pKa = 3.21GVADD1801 pKa = 3.73YY1802 pKa = 11.17TEE1804 pKa = 4.49FAGSDD1809 pKa = 3.98GKK1811 pKa = 10.27WSVRR1815 pKa = 11.84PDD1817 pKa = 3.17TPLANGTSVSVTSTDD1832 pKa = 3.2PVGNVSAAATTVVDD1846 pKa = 3.77NVAPVVTLITADD1858 pKa = 3.56NQTLTGTVDD1867 pKa = 3.43DD1868 pKa = 4.32STVTSVKK1875 pKa = 10.4VFDD1878 pKa = 3.54NTGNFVGTGTVTNGSYY1894 pKa = 11.02SLTLADD1900 pKa = 4.11PVAPGTVLTAKK1911 pKa = 9.71ATDD1914 pKa = 3.47PAGNVGSSSSLTIGSVSSDD1933 pKa = 3.05SDD1935 pKa = 3.36NSALATHH1942 pKa = 6.17QAAITAIATDD1952 pKa = 4.11TLPTSAVSTVQNNDD1966 pKa = 2.34FDD1968 pKa = 4.36TRR1970 pKa = 11.84DD1971 pKa = 3.48PTLTVSGTSNLEE1983 pKa = 3.89PAQEE1987 pKa = 3.53AAAASFIAMPLAMSLTSSNASSSTFSDD2014 pKa = 3.52VLQISTDD2021 pKa = 3.43GQNWSTVTPSANGAWTWTDD2040 pKa = 3.37PTAHH2044 pKa = 7.36DD2045 pKa = 4.72SNFTYY2050 pKa = 10.69YY2051 pKa = 10.82LRR2053 pKa = 11.84TLDD2056 pKa = 3.45SSGKK2060 pKa = 9.71VVYY2063 pKa = 7.62TTSKK2067 pKa = 10.94DD2068 pKa = 3.12ITIDD2072 pKa = 4.3LIAPSSLVMPTWNDD2086 pKa = 2.87AGITVTKK2093 pKa = 10.7DD2094 pKa = 2.6ADD2096 pKa = 3.59ATVALINDD2104 pKa = 3.73VNHH2107 pKa = 7.13DD2108 pKa = 3.52GVYY2111 pKa = 9.93EE2112 pKa = 3.99QGVDD2116 pKa = 3.25KK2117 pKa = 11.19VLAVADD2123 pKa = 3.98NSGTTATLSATLTAGQHH2140 pKa = 5.38YY2141 pKa = 10.56NLGLVQYY2148 pKa = 9.79DD2149 pKa = 3.38VAGNYY2154 pKa = 9.57SRR2156 pKa = 11.84LSNTVSYY2163 pKa = 7.61TAPPTSALNTAFQASVLPPQEE2184 pKa = 4.28VRR2186 pKa = 11.84DD2187 pKa = 4.03SAGMAVGFDD2196 pKa = 3.43AYY2198 pKa = 10.92GNLNILQRR2206 pKa = 11.84SSLITQSNSTNYY2218 pKa = 10.58ALSDD2222 pKa = 3.88VQQLPYY2228 pKa = 10.48LVRR2231 pKa = 11.84YY2232 pKa = 6.81TNQSTDD2238 pKa = 2.53SRR2240 pKa = 11.84NTINSSTIVDD2250 pKa = 3.91YY2251 pKa = 11.57NRR2253 pKa = 11.84DD2254 pKa = 3.47GYY2256 pKa = 11.56SDD2258 pKa = 4.04VLASDD2263 pKa = 4.98YY2264 pKa = 10.84IDD2266 pKa = 3.59SATTGVAVWTGSQSGYY2282 pKa = 10.26SLTRR2286 pKa = 11.84ATSSNNSAGGVMAYY2300 pKa = 10.36DD2301 pKa = 3.87KK2302 pKa = 11.49DD2303 pKa = 3.55GDD2305 pKa = 4.27GYY2307 pKa = 11.56VDD2309 pKa = 3.86AVLGNWGPQNGATNGTFIKK2328 pKa = 9.32NTNGVLSQDD2337 pKa = 3.78GTNGTAGLQNFQFEE2351 pKa = 4.67RR2352 pKa = 11.84EE2353 pKa = 4.09VSGVDD2358 pKa = 3.27LKK2360 pKa = 11.55GVGNIDD2366 pKa = 3.53VAAHH2370 pKa = 6.62TISNSASSNQYY2381 pKa = 11.23ALALIDD2387 pKa = 4.12NNGGTLSLGQNIDD2400 pKa = 3.08NVFNSRR2406 pKa = 11.84GIPTGTTGQKK2416 pKa = 10.39ALPSVWGAQSMVWADD2431 pKa = 3.74FANNGHH2437 pKa = 6.4MDD2439 pKa = 4.07LYY2441 pKa = 10.99LSQGGNTPSGSDD2453 pKa = 3.25NTDD2456 pKa = 2.45SRR2458 pKa = 11.84IYY2460 pKa = 9.95MNNNGVLSSTPTIIADD2476 pKa = 3.64DD2477 pKa = 4.01NLAGGAAIATDD2488 pKa = 3.65WNHH2491 pKa = 6.84DD2492 pKa = 3.36GKK2494 pKa = 10.75IDD2496 pKa = 3.85VIEE2499 pKa = 4.0IRR2501 pKa = 11.84TAIEE2505 pKa = 3.94NNVTNIPVTLLTNNGVNSNGTLNPFTVSNMTTIPRR2540 pKa = 11.84ANITGIAATDD2550 pKa = 3.89VNWDD2554 pKa = 3.53GAVDD2558 pKa = 4.32LLYY2561 pKa = 10.31STASGSDD2568 pKa = 3.27TQADD2572 pKa = 3.89KK2573 pKa = 11.3AGKK2576 pKa = 5.95TTEE2579 pKa = 4.04GNIYY2583 pKa = 10.54SIMNTNAVADD2593 pKa = 4.26GTSLHH2598 pKa = 6.93LKK2600 pKa = 10.1ILDD2603 pKa = 3.69QNGVNSYY2610 pKa = 10.53FGNTVQLFDD2619 pKa = 4.32SDD2621 pKa = 4.24GNLVSTQIINPQEE2634 pKa = 4.09GMWNNDD2640 pKa = 2.11SSGIVNFYY2648 pKa = 11.16NLDD2651 pKa = 3.88PNQHH2655 pKa = 5.68YY2656 pKa = 10.62SVALVRR2662 pKa = 11.84ATDD2665 pKa = 3.42GHH2667 pKa = 6.96SNDD2670 pKa = 2.84IGGRR2674 pKa = 11.84DD2675 pKa = 3.13SYY2677 pKa = 12.03GQFFTLSSVEE2687 pKa = 4.32DD2688 pKa = 3.63SSSVSDD2694 pKa = 3.87GGSEE2698 pKa = 3.84TGAASALNTVEE2709 pKa = 4.52NVEE2712 pKa = 4.25KK2713 pKa = 10.43SWNNLTPGAANHH2725 pKa = 6.61AYY2727 pKa = 10.12VLEE2730 pKa = 4.83GDD2732 pKa = 3.8TTASHH2737 pKa = 5.14TTGGIFTGTGYY2748 pKa = 11.12DD2749 pKa = 3.3DD2750 pKa = 3.76TFFAGAGNNSYY2761 pKa = 10.45IGGGGWGYY2769 pKa = 11.23NSAGEE2774 pKa = 4.22YY2775 pKa = 10.22VWMSSGGHH2783 pKa = 5.24NVVDD2787 pKa = 3.81YY2788 pKa = 10.85SGEE2791 pKa = 4.02SSAITVNMGNSVGIGTVSVSKK2812 pKa = 10.02TDD2814 pKa = 3.25GHH2816 pKa = 7.27DD2817 pKa = 3.51SLQSIQGIVGTNYY2830 pKa = 10.62DD2831 pKa = 4.54DD2832 pKa = 5.7SFTSDD2837 pKa = 2.61GKK2839 pKa = 11.23GDD2841 pKa = 3.69YY2842 pKa = 10.89FEE2844 pKa = 6.45GGGGNDD2850 pKa = 3.43TFNLAKK2856 pKa = 10.39GGHH2859 pKa = 5.99DD2860 pKa = 2.84TLMYY2864 pKa = 10.82KK2865 pKa = 10.72LLDD2868 pKa = 3.86NSDD2871 pKa = 3.59NTGGNGSDD2879 pKa = 3.56TVNDD2883 pKa = 3.49FHH2885 pKa = 8.75LGNTVNNPDD2894 pKa = 4.0ADD2896 pKa = 3.96IINIKK2901 pKa = 10.35DD2902 pKa = 3.82LLADD2906 pKa = 3.72YY2907 pKa = 10.79QGTANVFYY2915 pKa = 10.81DD2916 pKa = 3.34VTEE2919 pKa = 4.22NKK2921 pKa = 9.82FVMDD2925 pKa = 4.34KK2926 pKa = 10.97ASSGLDD2932 pKa = 3.12QFVSTSVHH2940 pKa = 6.05NGNTTISVDD2949 pKa = 3.23RR2950 pKa = 11.84DD2951 pKa = 3.5GANGNHH2957 pKa = 5.83QMTALVTLDD2966 pKa = 3.35NTQTDD2971 pKa = 4.3LATLIGNHH2979 pKa = 5.57QLIIAA2984 pKa = 4.8
Molecular weight: 306.83 kDa
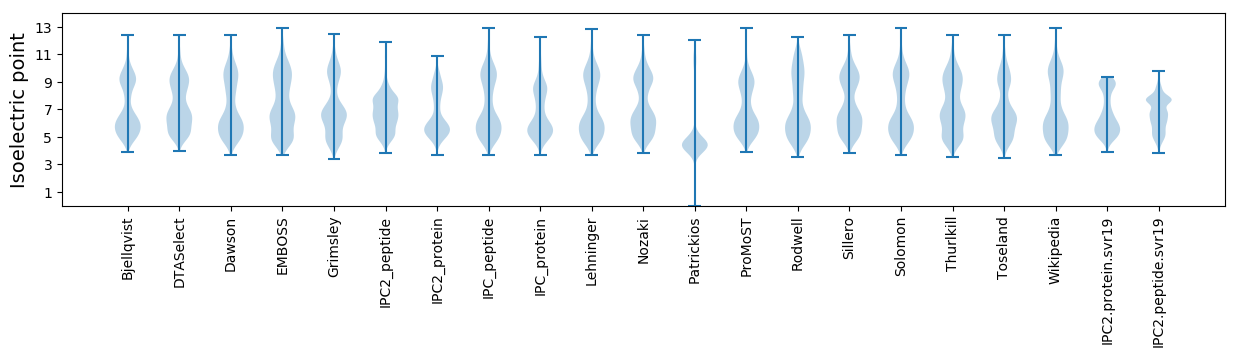
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5NR06|Q5NR06_ZYMMO Methionine biosynthesis protein MetW OS=Zymomonas mobilis subsp. mobilis (strain ATCC 31821 / ZM4 / CP4) OX=264203 GN=ZMO0224 PE=4 SV=1
MM1 pKa = 7.89RR2 pKa = 11.84FLSGLSSFYY11 pKa = 10.19KK12 pKa = 10.27AHH14 pKa = 7.14RR15 pKa = 11.84LLINLSICLPPMALLSLSIGSTHH38 pKa = 7.0IGIMALWQAVAGVGDD53 pKa = 3.78PTSRR57 pKa = 11.84LILGLLRR64 pKa = 11.84LPRR67 pKa = 11.84LCVGLAVGAILGMAGAVNQGYY88 pKa = 9.64LRR90 pKa = 11.84NPLADD95 pKa = 3.99PSLLGTSNAAALGAVLAFYY114 pKa = 10.55FGLSARR120 pKa = 11.84FPICLPLFAFIGALAGLLPLLWFVRR145 pKa = 11.84RR146 pKa = 11.84HH147 pKa = 6.07NDD149 pKa = 3.38PQSLILGGVAIAAFSGACISLALNLSPNPFAAMEE183 pKa = 4.01IMTWLMGAITDD194 pKa = 4.25RR195 pKa = 11.84SWNHH199 pKa = 4.22VWLLIAPFFLSLFLFRR215 pKa = 11.84RR216 pKa = 11.84TAPALDD222 pKa = 3.72ALILGDD228 pKa = 5.35DD229 pKa = 3.88VAASLGFDD237 pKa = 4.19LARR240 pKa = 11.84LRR242 pKa = 11.84WLIMAALAIGVGGAVAVSGAIGFVGLIVPHH272 pKa = 6.93ILRR275 pKa = 11.84PFTRR279 pKa = 11.84QLPSEE284 pKa = 4.55LLWPSAIGGALLVTLADD301 pKa = 3.21ILVRR305 pKa = 11.84LLPTQNEE312 pKa = 4.17LQLGVVTAFIGVPIFFRR329 pKa = 11.84QAIKK333 pKa = 10.75GVGAMKK339 pKa = 10.53SLL341 pKa = 3.62
MM1 pKa = 7.89RR2 pKa = 11.84FLSGLSSFYY11 pKa = 10.19KK12 pKa = 10.27AHH14 pKa = 7.14RR15 pKa = 11.84LLINLSICLPPMALLSLSIGSTHH38 pKa = 7.0IGIMALWQAVAGVGDD53 pKa = 3.78PTSRR57 pKa = 11.84LILGLLRR64 pKa = 11.84LPRR67 pKa = 11.84LCVGLAVGAILGMAGAVNQGYY88 pKa = 9.64LRR90 pKa = 11.84NPLADD95 pKa = 3.99PSLLGTSNAAALGAVLAFYY114 pKa = 10.55FGLSARR120 pKa = 11.84FPICLPLFAFIGALAGLLPLLWFVRR145 pKa = 11.84RR146 pKa = 11.84HH147 pKa = 6.07NDD149 pKa = 3.38PQSLILGGVAIAAFSGACISLALNLSPNPFAAMEE183 pKa = 4.01IMTWLMGAITDD194 pKa = 4.25RR195 pKa = 11.84SWNHH199 pKa = 4.22VWLLIAPFFLSLFLFRR215 pKa = 11.84RR216 pKa = 11.84TAPALDD222 pKa = 3.72ALILGDD228 pKa = 5.35DD229 pKa = 3.88VAASLGFDD237 pKa = 4.19LARR240 pKa = 11.84LRR242 pKa = 11.84WLIMAALAIGVGGAVAVSGAIGFVGLIVPHH272 pKa = 6.93ILRR275 pKa = 11.84PFTRR279 pKa = 11.84QLPSEE284 pKa = 4.55LLWPSAIGGALLVTLADD301 pKa = 3.21ILVRR305 pKa = 11.84LLPTQNEE312 pKa = 4.17LQLGVVTAFIGVPIFFRR329 pKa = 11.84QAIKK333 pKa = 10.75GVGAMKK339 pKa = 10.53SLL341 pKa = 3.62
Molecular weight: 36.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
599273 |
30 |
2984 |
336.9 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.183 ± 0.072 | 0.835 ± 0.019 |
5.722 ± 0.044 | 5.666 ± 0.065 |
3.887 ± 0.041 | 7.461 ± 0.067 |
2.335 ± 0.026 | 6.531 ± 0.047 |
5.058 ± 0.048 | 9.991 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.422 ± 0.025 | 3.787 ± 0.049 |
4.737 ± 0.033 | 3.807 ± 0.033 |
5.694 ± 0.054 | 6.668 ± 0.055 |
5.059 ± 0.054 | 6.225 ± 0.049 |
1.277 ± 0.022 | 2.651 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
