
Vibrio genomosp. F10
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
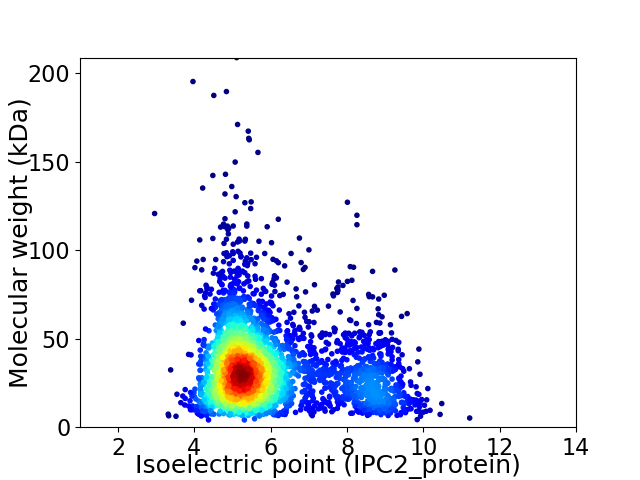
Virtual 2D-PAGE plot for 3193 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
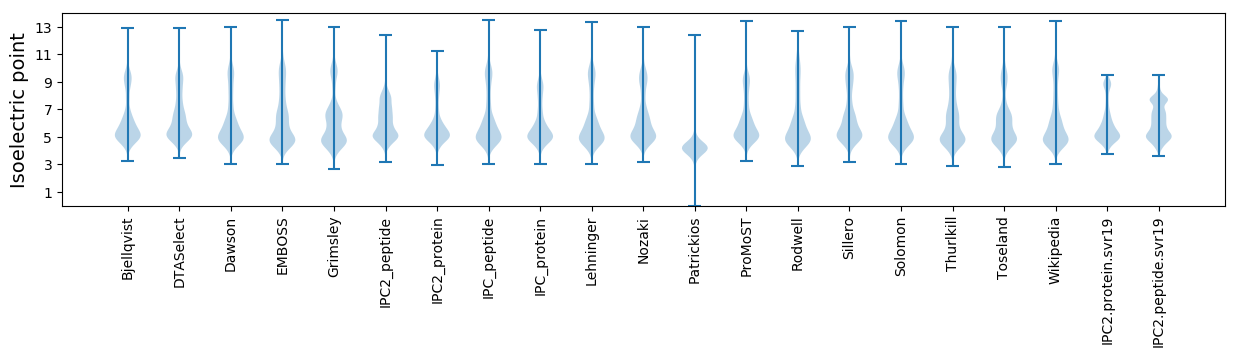
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B9R1J0|A0A1B9R1J0_9VIBR Uncharacterized protein OS=Vibrio genomosp. F10 OX=723171 GN=A6E14_06865 PE=4 SV=1
MM1 pKa = 7.35FNIKK5 pKa = 9.63SLPMAICLGLAALSGSAASASAMAGEE31 pKa = 4.58MVNPDD36 pKa = 3.05GNVVVGYY43 pKa = 6.91WHH45 pKa = 6.68NWCDD49 pKa = 3.29GAGYY53 pKa = 10.39KK54 pKa = 10.38GGNSPCMSLDD64 pKa = 5.11DD65 pKa = 4.69INPMYY70 pKa = 10.82NVVDD74 pKa = 3.7ISFMKK79 pKa = 10.7VYY81 pKa = 9.18DD82 pKa = 3.84TAEE85 pKa = 3.76GRR87 pKa = 11.84IPTFKK92 pKa = 10.76LDD94 pKa = 3.54PTVGLSEE101 pKa = 4.36QEE103 pKa = 4.48FINQIEE109 pKa = 4.22QLNQQGRR116 pKa = 11.84SVLLALGGADD126 pKa = 3.84AHH128 pKa = 7.43VEE130 pKa = 4.0LQKK133 pKa = 11.17GDD135 pKa = 3.51EE136 pKa = 3.97QAFAEE141 pKa = 4.64EE142 pKa = 4.77IIRR145 pKa = 11.84LTEE148 pKa = 3.62QYY150 pKa = 11.18GFDD153 pKa = 3.96GLDD156 pKa = 3.07IDD158 pKa = 5.36LEE160 pKa = 4.37QAAVTAADD168 pKa = 3.71NQIVIPAALKK178 pKa = 10.05IVKK181 pKa = 9.44DD182 pKa = 3.73HH183 pKa = 6.42YY184 pKa = 11.11AAEE187 pKa = 4.54GKK189 pKa = 10.17NFLITMAPEE198 pKa = 4.36FPYY201 pKa = 9.54LTQGGKK207 pKa = 9.32YY208 pKa = 9.76VPYY211 pKa = 9.86IDD213 pKa = 5.19NLAGYY218 pKa = 8.89YY219 pKa = 10.41DD220 pKa = 4.32WINPQFYY227 pKa = 10.24NQGGDD232 pKa = 4.12GIWVDD237 pKa = 3.71SVGWIAQNNDD247 pKa = 2.84ALKK250 pKa = 10.7QEE252 pKa = 4.84FIYY255 pKa = 10.92YY256 pKa = 9.95IADD259 pKa = 3.49SLSNGTRR266 pKa = 11.84GFHH269 pKa = 6.38QIPHH273 pKa = 6.11NKK275 pKa = 9.59LVFGIPANNDD285 pKa = 2.95AAATGYY291 pKa = 8.94VQNPQDD297 pKa = 4.1LYY299 pKa = 11.63DD300 pKa = 4.74AFEE303 pKa = 4.0QLNQQGQPLRR313 pKa = 11.84GVMTWSVNWDD323 pKa = 3.25MGTDD327 pKa = 3.34AGGRR331 pKa = 11.84AYY333 pKa = 10.74NEE335 pKa = 3.68QFIRR339 pKa = 11.84DD340 pKa = 4.1YY341 pKa = 11.24GPYY344 pKa = 9.56IHH346 pKa = 6.94GQTPPPITSNEE357 pKa = 3.94PVLKK361 pKa = 10.82GIDD364 pKa = 3.73DD365 pKa = 3.83ARR367 pKa = 11.84VAHH370 pKa = 6.1QSYY373 pKa = 9.42FDD375 pKa = 3.82PMSDD379 pKa = 3.07VTAMDD384 pKa = 5.38DD385 pKa = 3.49EE386 pKa = 7.32DD387 pKa = 6.6GDD389 pKa = 4.2LTHH392 pKa = 7.33HH393 pKa = 6.2ITVEE397 pKa = 4.26GQVNTSSVGTYY408 pKa = 10.5ALTYY412 pKa = 10.36LVEE415 pKa = 5.81DD416 pKa = 3.53SDD418 pKa = 4.75GNQTSKK424 pKa = 10.44VRR426 pKa = 11.84SVDD429 pKa = 3.62VYY431 pKa = 10.51SSKK434 pKa = 10.92PIFKK438 pKa = 10.23GVSDD442 pKa = 3.55TSIQFGSEE450 pKa = 3.73FDD452 pKa = 3.67SLAGVSASDD461 pKa = 4.53EE462 pKa = 4.18EE463 pKa = 6.15DD464 pKa = 3.49GDD466 pKa = 3.8LTSRR470 pKa = 11.84IEE472 pKa = 3.79IYY474 pKa = 11.17GDD476 pKa = 3.44VEE478 pKa = 4.38TNRR481 pKa = 11.84LGSQTLLYY489 pKa = 10.77SVTDD493 pKa = 3.59SANQSTTVEE502 pKa = 4.15RR503 pKa = 11.84TIHH506 pKa = 5.97VVDD509 pKa = 5.05EE510 pKa = 4.68SSCAAPWQTNNVYY523 pKa = 10.58EE524 pKa = 4.56GGDD527 pKa = 3.31RR528 pKa = 11.84VLHH531 pKa = 6.85DD532 pKa = 3.39GTVYY536 pKa = 10.03EE537 pKa = 4.29AAWWTRR543 pKa = 11.84GEE545 pKa = 4.29QPGSTGQWGVWRR557 pKa = 11.84EE558 pKa = 4.05VTDD561 pKa = 3.7TGCDD565 pKa = 3.32GTNNGDD571 pKa = 3.73NGAGDD576 pKa = 4.54DD577 pKa = 5.13NNNGDD582 pKa = 4.29NNNSGDD588 pKa = 3.86TDD590 pKa = 3.69NGSDD594 pKa = 3.36TGNEE598 pKa = 4.01NGDD601 pKa = 3.97SNGGNGNTDD610 pKa = 3.82NVTEE614 pKa = 5.07WIPGQTQASNGDD626 pKa = 3.52MVSYY630 pKa = 10.5QGSCFVAKK638 pKa = 10.4NNPGVWEE645 pKa = 4.61TPTQSATWFWDD656 pKa = 3.75EE657 pKa = 4.36VSCPP661 pKa = 3.51
MM1 pKa = 7.35FNIKK5 pKa = 9.63SLPMAICLGLAALSGSAASASAMAGEE31 pKa = 4.58MVNPDD36 pKa = 3.05GNVVVGYY43 pKa = 6.91WHH45 pKa = 6.68NWCDD49 pKa = 3.29GAGYY53 pKa = 10.39KK54 pKa = 10.38GGNSPCMSLDD64 pKa = 5.11DD65 pKa = 4.69INPMYY70 pKa = 10.82NVVDD74 pKa = 3.7ISFMKK79 pKa = 10.7VYY81 pKa = 9.18DD82 pKa = 3.84TAEE85 pKa = 3.76GRR87 pKa = 11.84IPTFKK92 pKa = 10.76LDD94 pKa = 3.54PTVGLSEE101 pKa = 4.36QEE103 pKa = 4.48FINQIEE109 pKa = 4.22QLNQQGRR116 pKa = 11.84SVLLALGGADD126 pKa = 3.84AHH128 pKa = 7.43VEE130 pKa = 4.0LQKK133 pKa = 11.17GDD135 pKa = 3.51EE136 pKa = 3.97QAFAEE141 pKa = 4.64EE142 pKa = 4.77IIRR145 pKa = 11.84LTEE148 pKa = 3.62QYY150 pKa = 11.18GFDD153 pKa = 3.96GLDD156 pKa = 3.07IDD158 pKa = 5.36LEE160 pKa = 4.37QAAVTAADD168 pKa = 3.71NQIVIPAALKK178 pKa = 10.05IVKK181 pKa = 9.44DD182 pKa = 3.73HH183 pKa = 6.42YY184 pKa = 11.11AAEE187 pKa = 4.54GKK189 pKa = 10.17NFLITMAPEE198 pKa = 4.36FPYY201 pKa = 9.54LTQGGKK207 pKa = 9.32YY208 pKa = 9.76VPYY211 pKa = 9.86IDD213 pKa = 5.19NLAGYY218 pKa = 8.89YY219 pKa = 10.41DD220 pKa = 4.32WINPQFYY227 pKa = 10.24NQGGDD232 pKa = 4.12GIWVDD237 pKa = 3.71SVGWIAQNNDD247 pKa = 2.84ALKK250 pKa = 10.7QEE252 pKa = 4.84FIYY255 pKa = 10.92YY256 pKa = 9.95IADD259 pKa = 3.49SLSNGTRR266 pKa = 11.84GFHH269 pKa = 6.38QIPHH273 pKa = 6.11NKK275 pKa = 9.59LVFGIPANNDD285 pKa = 2.95AAATGYY291 pKa = 8.94VQNPQDD297 pKa = 4.1LYY299 pKa = 11.63DD300 pKa = 4.74AFEE303 pKa = 4.0QLNQQGQPLRR313 pKa = 11.84GVMTWSVNWDD323 pKa = 3.25MGTDD327 pKa = 3.34AGGRR331 pKa = 11.84AYY333 pKa = 10.74NEE335 pKa = 3.68QFIRR339 pKa = 11.84DD340 pKa = 4.1YY341 pKa = 11.24GPYY344 pKa = 9.56IHH346 pKa = 6.94GQTPPPITSNEE357 pKa = 3.94PVLKK361 pKa = 10.82GIDD364 pKa = 3.73DD365 pKa = 3.83ARR367 pKa = 11.84VAHH370 pKa = 6.1QSYY373 pKa = 9.42FDD375 pKa = 3.82PMSDD379 pKa = 3.07VTAMDD384 pKa = 5.38DD385 pKa = 3.49EE386 pKa = 7.32DD387 pKa = 6.6GDD389 pKa = 4.2LTHH392 pKa = 7.33HH393 pKa = 6.2ITVEE397 pKa = 4.26GQVNTSSVGTYY408 pKa = 10.5ALTYY412 pKa = 10.36LVEE415 pKa = 5.81DD416 pKa = 3.53SDD418 pKa = 4.75GNQTSKK424 pKa = 10.44VRR426 pKa = 11.84SVDD429 pKa = 3.62VYY431 pKa = 10.51SSKK434 pKa = 10.92PIFKK438 pKa = 10.23GVSDD442 pKa = 3.55TSIQFGSEE450 pKa = 3.73FDD452 pKa = 3.67SLAGVSASDD461 pKa = 4.53EE462 pKa = 4.18EE463 pKa = 6.15DD464 pKa = 3.49GDD466 pKa = 3.8LTSRR470 pKa = 11.84IEE472 pKa = 3.79IYY474 pKa = 11.17GDD476 pKa = 3.44VEE478 pKa = 4.38TNRR481 pKa = 11.84LGSQTLLYY489 pKa = 10.77SVTDD493 pKa = 3.59SANQSTTVEE502 pKa = 4.15RR503 pKa = 11.84TIHH506 pKa = 5.97VVDD509 pKa = 5.05EE510 pKa = 4.68SSCAAPWQTNNVYY523 pKa = 10.58EE524 pKa = 4.56GGDD527 pKa = 3.31RR528 pKa = 11.84VLHH531 pKa = 6.85DD532 pKa = 3.39GTVYY536 pKa = 10.03EE537 pKa = 4.29AAWWTRR543 pKa = 11.84GEE545 pKa = 4.29QPGSTGQWGVWRR557 pKa = 11.84EE558 pKa = 4.05VTDD561 pKa = 3.7TGCDD565 pKa = 3.32GTNNGDD571 pKa = 3.73NGAGDD576 pKa = 4.54DD577 pKa = 5.13NNNGDD582 pKa = 4.29NNNSGDD588 pKa = 3.86TDD590 pKa = 3.69NGSDD594 pKa = 3.36TGNEE598 pKa = 4.01NGDD601 pKa = 3.97SNGGNGNTDD610 pKa = 3.82NVTEE614 pKa = 5.07WIPGQTQASNGDD626 pKa = 3.52MVSYY630 pKa = 10.5QGSCFVAKK638 pKa = 10.4NNPGVWEE645 pKa = 4.61TPTQSATWFWDD656 pKa = 3.75EE657 pKa = 4.36VSCPP661 pKa = 3.51
Molecular weight: 71.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9R0H4|A0A1B9R0H4_9VIBR PBPb domain-containing protein OS=Vibrio genomosp. F10 OX=723171 GN=A6E14_01210 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.5RR3 pKa = 11.84TFQPTVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84ATINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
MM1 pKa = 7.45KK2 pKa = 9.5RR3 pKa = 11.84TFQPTVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84ATINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1029883 |
35 |
1908 |
322.5 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.446 ± 0.043 | 0.994 ± 0.013 |
5.572 ± 0.038 | 6.376 ± 0.041 |
4.086 ± 0.03 | 6.871 ± 0.043 |
2.283 ± 0.022 | 6.514 ± 0.034 |
5.236 ± 0.034 | 10.357 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.021 | 4.196 ± 0.03 |
3.855 ± 0.027 | 4.508 ± 0.031 |
4.478 ± 0.033 | 6.871 ± 0.039 |
5.422 ± 0.031 | 7.129 ± 0.038 |
1.221 ± 0.016 | 2.881 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
