
Sulfolobus islandicus filamentous virus 2
Taxonomy: Viruses; Adnaviria; Zilligvirae; Taleaviricota; Tokiviricetes; Ligamenvirales; Lipothrixviridae; Betalipothrixvirus; Sulfolobus islandicus filamentous virus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
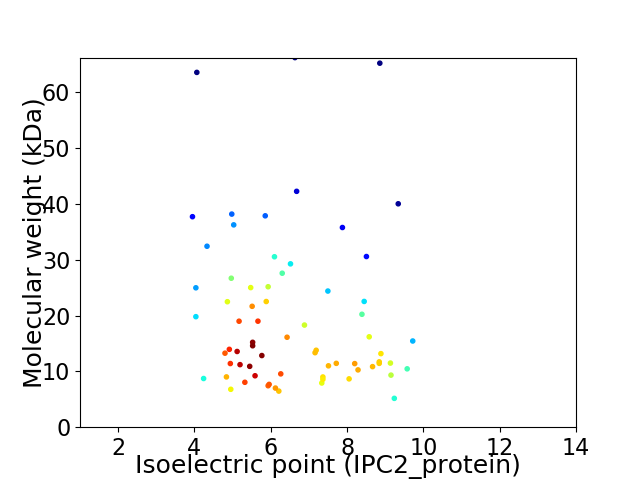
Virtual 2D-PAGE plot for 69 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8BJA7|A0A1D8BJA7_SIFV SIFV.gp60-like protein OS=Sulfolobus islandicus filamentous virus 2 OX=1902331 GN=SIFV2_gp55 PE=4 SV=1
MM1 pKa = 7.77SFLLNLGQAFFSGLSSIATGVTSDD25 pKa = 3.51VTSVANGINYY35 pKa = 9.11AFNVLAKK42 pKa = 9.87FLQKK46 pKa = 10.56LPSEE50 pKa = 4.11IATFFQSIPGALVAFAHH67 pKa = 7.1AFGTYY72 pKa = 9.48LWDD75 pKa = 3.56GMQRR79 pKa = 11.84IASGFTTIMAPIEE92 pKa = 4.47RR93 pKa = 11.84GLEE96 pKa = 4.05TLGTTVINALTVVWNDD112 pKa = 2.43IKK114 pKa = 11.33AFASAIYY121 pKa = 10.3NSIVSMINNIVTFVQPFINDD141 pKa = 3.8LKK143 pKa = 10.65NAISFAYY150 pKa = 10.29NFLLNVINDD159 pKa = 3.49VYY161 pKa = 11.42SAFTIISNFFLDD173 pKa = 5.26LPTFFQNASNYY184 pKa = 10.02LNSLFTDD191 pKa = 4.43PGNQPNLLTMVPNLVASEE209 pKa = 4.07VSRR212 pKa = 11.84IAGAFPDD219 pKa = 4.22VIAYY223 pKa = 6.7NTFMEE228 pKa = 4.74VLPKK232 pKa = 9.45MVSGIASSPIFGNTVKK248 pKa = 10.84GMFAKK253 pKa = 10.37ALLMAGSPILSALLSEE269 pKa = 4.33LTKK272 pKa = 11.29ALMQSLFTSTQTTQTTQRR290 pKa = 11.84PSQPQIQPPSAPSTQLQQRR309 pKa = 11.84STQQTSLSDD318 pKa = 3.65LQNLQTQQLTPTQIEE333 pKa = 4.49VQLEE337 pKa = 4.24RR338 pKa = 11.84PNVTGVFTQDD348 pKa = 3.36VIGMGTASGGTAKK361 pKa = 10.53LVSGYY366 pKa = 10.59INFQNSIKK374 pKa = 10.4QFEE377 pKa = 4.59DD378 pKa = 3.38VFDD381 pKa = 3.53ITASFFMEE389 pKa = 5.74LIQSLQTEE397 pKa = 5.28FSQDD401 pKa = 2.89MAVNVSLTLLEE412 pKa = 4.75NIYY415 pKa = 10.33QVSQAIEE422 pKa = 4.28IIPSVDD428 pKa = 2.53ATTIVLPPGISLCNPSQAPSPSEE451 pKa = 4.02STSTSTNSIDD461 pKa = 3.18VTTTVDD467 pKa = 3.36VTEE470 pKa = 4.25QLCIPPYY477 pKa = 10.77DD478 pKa = 4.12LLADD482 pKa = 4.18GLTTQFAVIALLHH495 pKa = 6.18SNISDD500 pKa = 3.32VMLAITQLIYY510 pKa = 10.82GITFSNLISDD520 pKa = 4.35ILSASTQLVYY530 pKa = 10.89SITSNISTSDD540 pKa = 3.43TVSVSASLTPSPPPSEE556 pKa = 4.37SYY558 pKa = 10.01TYY560 pKa = 10.2YY561 pKa = 10.91VPIDD565 pKa = 3.49VAVTYY570 pKa = 7.72TIQIASSTTYY580 pKa = 10.69SGTLSYY586 pKa = 10.22TVSTSS591 pKa = 2.73
MM1 pKa = 7.77SFLLNLGQAFFSGLSSIATGVTSDD25 pKa = 3.51VTSVANGINYY35 pKa = 9.11AFNVLAKK42 pKa = 9.87FLQKK46 pKa = 10.56LPSEE50 pKa = 4.11IATFFQSIPGALVAFAHH67 pKa = 7.1AFGTYY72 pKa = 9.48LWDD75 pKa = 3.56GMQRR79 pKa = 11.84IASGFTTIMAPIEE92 pKa = 4.47RR93 pKa = 11.84GLEE96 pKa = 4.05TLGTTVINALTVVWNDD112 pKa = 2.43IKK114 pKa = 11.33AFASAIYY121 pKa = 10.3NSIVSMINNIVTFVQPFINDD141 pKa = 3.8LKK143 pKa = 10.65NAISFAYY150 pKa = 10.29NFLLNVINDD159 pKa = 3.49VYY161 pKa = 11.42SAFTIISNFFLDD173 pKa = 5.26LPTFFQNASNYY184 pKa = 10.02LNSLFTDD191 pKa = 4.43PGNQPNLLTMVPNLVASEE209 pKa = 4.07VSRR212 pKa = 11.84IAGAFPDD219 pKa = 4.22VIAYY223 pKa = 6.7NTFMEE228 pKa = 4.74VLPKK232 pKa = 9.45MVSGIASSPIFGNTVKK248 pKa = 10.84GMFAKK253 pKa = 10.37ALLMAGSPILSALLSEE269 pKa = 4.33LTKK272 pKa = 11.29ALMQSLFTSTQTTQTTQRR290 pKa = 11.84PSQPQIQPPSAPSTQLQQRR309 pKa = 11.84STQQTSLSDD318 pKa = 3.65LQNLQTQQLTPTQIEE333 pKa = 4.49VQLEE337 pKa = 4.24RR338 pKa = 11.84PNVTGVFTQDD348 pKa = 3.36VIGMGTASGGTAKK361 pKa = 10.53LVSGYY366 pKa = 10.59INFQNSIKK374 pKa = 10.4QFEE377 pKa = 4.59DD378 pKa = 3.38VFDD381 pKa = 3.53ITASFFMEE389 pKa = 5.74LIQSLQTEE397 pKa = 5.28FSQDD401 pKa = 2.89MAVNVSLTLLEE412 pKa = 4.75NIYY415 pKa = 10.33QVSQAIEE422 pKa = 4.28IIPSVDD428 pKa = 2.53ATTIVLPPGISLCNPSQAPSPSEE451 pKa = 4.02STSTSTNSIDD461 pKa = 3.18VTTTVDD467 pKa = 3.36VTEE470 pKa = 4.25QLCIPPYY477 pKa = 10.77DD478 pKa = 4.12LLADD482 pKa = 4.18GLTTQFAVIALLHH495 pKa = 6.18SNISDD500 pKa = 3.32VMLAITQLIYY510 pKa = 10.82GITFSNLISDD520 pKa = 4.35ILSASTQLVYY530 pKa = 10.89SITSNISTSDD540 pKa = 3.43TVSVSASLTPSPPPSEE556 pKa = 4.37SYY558 pKa = 10.01TYY560 pKa = 10.2YY561 pKa = 10.91VPIDD565 pKa = 3.49VAVTYY570 pKa = 7.72TIQIASSTTYY580 pKa = 10.69SGTLSYY586 pKa = 10.22TVSTSS591 pKa = 2.73
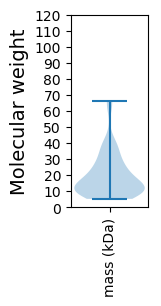
Molecular weight: 63.57 kDa
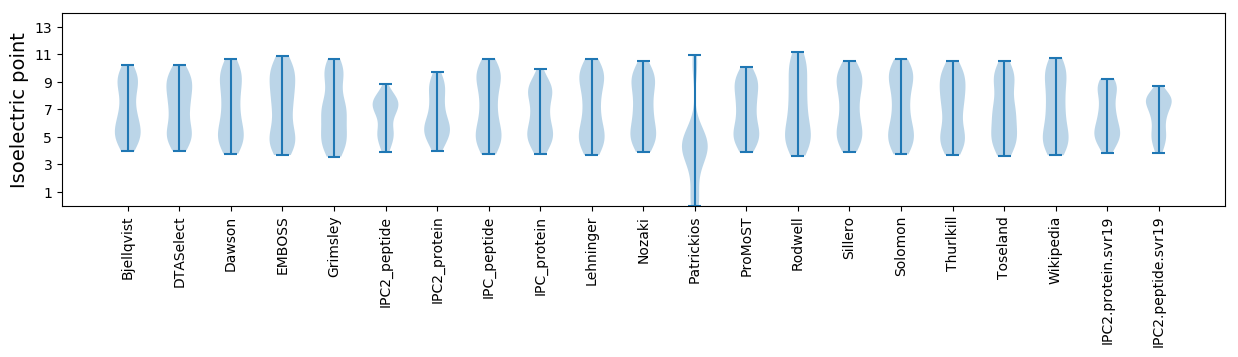
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8BJA2|A0A1D8BJA2_SIFV SIFV.gp55-like protein OS=Sulfolobus islandicus filamentous virus 2 OX=1902331 GN=SIFV2_gp50 PE=4 SV=1
MM1 pKa = 6.7VHH3 pKa = 7.08ALLSHH8 pKa = 6.73VITYY12 pKa = 7.67TKK14 pKa = 10.46FIVLVVKK21 pKa = 9.86LLIMSAIAGVIAGAFGGGIGGVGNAIGTIIGDD53 pKa = 3.99LEE55 pKa = 3.76RR56 pKa = 11.84AIARR60 pKa = 11.84FGGSIVNAFKK70 pKa = 10.39TVIDD74 pKa = 4.2KK75 pKa = 10.84VLSLAVRR82 pKa = 11.84IGRR85 pKa = 11.84IIEE88 pKa = 4.01KK89 pKa = 8.88YY90 pKa = 9.9FRR92 pKa = 11.84IAVHH96 pKa = 6.0YY97 pKa = 9.95IVLFLRR103 pKa = 11.84LAYY106 pKa = 9.74KK107 pKa = 10.54YY108 pKa = 9.04MYY110 pKa = 10.6SFYY113 pKa = 10.9TEE115 pKa = 3.97FQKK118 pKa = 11.18DD119 pKa = 3.44PWRR122 pKa = 11.84SLQFIGSMAILLNNSLFPP140 pKa = 4.81
MM1 pKa = 6.7VHH3 pKa = 7.08ALLSHH8 pKa = 6.73VITYY12 pKa = 7.67TKK14 pKa = 10.46FIVLVVKK21 pKa = 9.86LLIMSAIAGVIAGAFGGGIGGVGNAIGTIIGDD53 pKa = 3.99LEE55 pKa = 3.76RR56 pKa = 11.84AIARR60 pKa = 11.84FGGSIVNAFKK70 pKa = 10.39TVIDD74 pKa = 4.2KK75 pKa = 10.84VLSLAVRR82 pKa = 11.84IGRR85 pKa = 11.84IIEE88 pKa = 4.01KK89 pKa = 8.88YY90 pKa = 9.9FRR92 pKa = 11.84IAVHH96 pKa = 6.0YY97 pKa = 9.95IVLFLRR103 pKa = 11.84LAYY106 pKa = 9.74KK107 pKa = 10.54YY108 pKa = 9.04MYY110 pKa = 10.6SFYY113 pKa = 10.9TEE115 pKa = 3.97FQKK118 pKa = 11.18DD119 pKa = 3.44PWRR122 pKa = 11.84SLQFIGSMAILLNNSLFPP140 pKa = 4.81
Molecular weight: 15.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11868 |
44 |
591 |
172.0 |
19.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.87 ± 0.304 | 1.07 ± 0.133 |
4.491 ± 0.267 | 6.741 ± 0.504 |
4.66 ± 0.199 | 4.971 ± 0.23 |
1.491 ± 0.148 | 9.319 ± 0.293 |
7.895 ± 0.646 | 9.378 ± 0.229 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.134 | 6.041 ± 0.286 |
3.556 ± 0.214 | 3.446 ± 0.297 |
3.278 ± 0.279 | 6.48 ± 0.508 |
6.277 ± 0.531 | 7.095 ± 0.271 |
0.868 ± 0.114 | 5.747 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
