
bacterium HR17
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2781 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
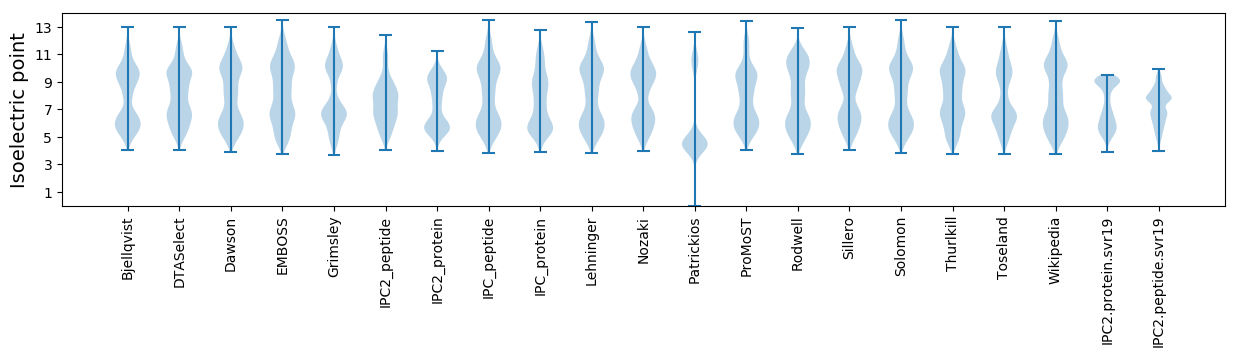
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5XAE8|A0A2H5XAE8_9BACT Uncharacterized protein OS=bacterium HR17 OX=2035412 GN=HRbin17_00659 PE=4 SV=1
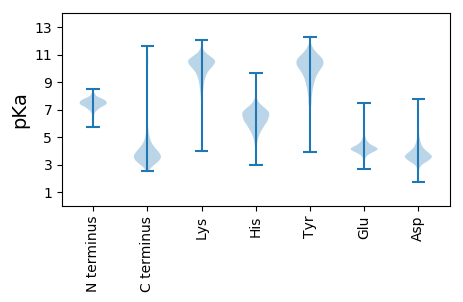
MM1 pKa = 7.73AKK3 pKa = 10.44GSLSVMLCLVGGFKK17 pKa = 10.44EE18 pKa = 4.66SEE20 pKa = 4.28GSQPSYY26 pKa = 10.35YY27 pKa = 9.74PPYY30 pKa = 9.88PPPQHH35 pKa = 4.81QHH37 pKa = 4.22NWTAGSSIPKK47 pKa = 8.72PIVMLFYY54 pKa = 10.5PGTSYY59 pKa = 10.48TLTHH63 pKa = 6.23LRR65 pKa = 11.84CYY67 pKa = 10.27EE68 pKa = 3.8IAFLADD74 pKa = 3.18VASATDD80 pKa = 3.68WDD82 pKa = 3.7QCIAPGCPYY91 pKa = 10.32PNGWAADD98 pKa = 3.65SKK100 pKa = 10.86VRR102 pKa = 11.84VMWSDD107 pKa = 3.12NGAGGQFGTLDD118 pKa = 3.55SNGNFIPLDD127 pKa = 3.59PVVNAQQITHH137 pKa = 5.98YY138 pKa = 9.63RR139 pKa = 11.84APWDD143 pKa = 3.45RR144 pKa = 11.84PGIVHH149 pKa = 6.27ITLVVDD155 pKa = 5.46DD156 pKa = 4.89EE157 pKa = 4.41GLYY160 pKa = 10.81YY161 pKa = 10.41DD162 pKa = 5.7DD163 pKa = 5.64PPVSNNSDD171 pKa = 3.45NGNVTVWEE179 pKa = 4.5FWITPCPYY187 pKa = 9.62NWRR190 pKa = 11.84PIPNTTGPSFIAWVEE205 pKa = 3.97PEE207 pKa = 4.13VDD209 pKa = 3.47HH210 pKa = 7.1LGQPMSGIMTFNLISSTEE228 pKa = 3.84PGEE231 pKa = 4.2CLNSHH236 pKa = 6.87FSCPYY241 pKa = 11.15DD242 pKa = 3.37MDD244 pKa = 3.48IVQTEE249 pKa = 3.97EE250 pKa = 4.64GEE252 pKa = 4.4TFEE255 pKa = 5.31VNDD258 pKa = 4.05IHH260 pKa = 8.16DD261 pKa = 4.85HH262 pKa = 6.8DD263 pKa = 6.22ADD265 pKa = 3.87LQFPPEE271 pKa = 4.1LEE273 pKa = 3.92GMNVYY278 pKa = 10.28GSQEE282 pKa = 3.78EE283 pKa = 4.65DD284 pKa = 3.27EE285 pKa = 4.77EE286 pKa = 5.28GPYY289 pKa = 8.64THH291 pKa = 6.96NFTVAVTQQPTTVAEE306 pKa = 4.05VRR308 pKa = 11.84ILCFDD313 pKa = 3.53GGGYY317 pKa = 10.75GFLLAGIDD325 pKa = 3.85GFPPGSATARR335 pKa = 11.84RR336 pKa = 11.84TDD338 pKa = 3.13TGVIGSVEE346 pKa = 3.78IPYY349 pKa = 9.99DD350 pKa = 3.56ANGNFIADD358 pKa = 3.19VWEE361 pKa = 4.45ANMGIYY367 pKa = 9.77PADD370 pKa = 3.51SLGDD374 pKa = 3.61FDD376 pKa = 4.64NQPVGDD382 pKa = 4.03GTLGDD387 pKa = 3.74GLSVYY392 pKa = 10.33EE393 pKa = 4.4EE394 pKa = 4.01YY395 pKa = 10.58RR396 pKa = 11.84GLRR399 pKa = 11.84VQGIWTVFNPRR410 pKa = 11.84LKK412 pKa = 11.26DD413 pKa = 3.24MFVMNFGAGQEE424 pKa = 4.2DD425 pKa = 5.53PYY427 pKa = 11.76VPATIPNEE435 pKa = 4.47AITSQDD441 pKa = 3.13GFPGQGMPLLWLLNARR457 pKa = 11.84EE458 pKa = 4.07GLGYY462 pKa = 9.21PLRR465 pKa = 11.84VVNYY469 pKa = 8.78LNHH472 pKa = 6.15YY473 pKa = 8.8AHH475 pKa = 6.86IRR477 pKa = 11.84DD478 pKa = 3.61VYY480 pKa = 10.78AAIVVPGDD488 pKa = 3.12IEE490 pKa = 4.92GYY492 pKa = 10.27QDD494 pKa = 4.45ADD496 pKa = 3.71NDD498 pKa = 4.3GQPDD502 pKa = 3.8DD503 pKa = 4.13TNGDD507 pKa = 3.79GVIDD511 pKa = 4.71DD512 pKa = 4.52SDD514 pKa = 5.4LIVDD518 pKa = 3.55VRR520 pKa = 11.84AYY522 pKa = 10.44GFTYY526 pKa = 10.47GPIWDD531 pKa = 3.99NAGGPIAPPGDD542 pKa = 3.17WGYY545 pKa = 10.61IPQFWGPPVIEE556 pKa = 4.15VDD558 pKa = 2.92VDD560 pKa = 3.3YY561 pKa = 11.39LRR563 pKa = 11.84NEE565 pKa = 4.07RR566 pKa = 11.84LDD568 pKa = 3.93PNTNPNGLTEE578 pKa = 4.26LQVLTWVVGHH588 pKa = 6.03EE589 pKa = 4.32LGHH592 pKa = 5.27TVLWHH597 pKa = 6.34RR598 pKa = 11.84NDD600 pKa = 3.33LGGYY604 pKa = 6.92GHH606 pKa = 7.28HH607 pKa = 6.96NGFDD611 pKa = 3.42AAGNPITTYY620 pKa = 11.25DD621 pKa = 3.69CLIWQWMDD629 pKa = 2.56WRR631 pKa = 11.84ANPPTEE637 pKa = 4.06FCAVNPGDD645 pKa = 3.55QTRR648 pKa = 11.84WKK650 pKa = 10.24INPP653 pKa = 3.5
MM1 pKa = 7.73AKK3 pKa = 10.44GSLSVMLCLVGGFKK17 pKa = 10.44EE18 pKa = 4.66SEE20 pKa = 4.28GSQPSYY26 pKa = 10.35YY27 pKa = 9.74PPYY30 pKa = 9.88PPPQHH35 pKa = 4.81QHH37 pKa = 4.22NWTAGSSIPKK47 pKa = 8.72PIVMLFYY54 pKa = 10.5PGTSYY59 pKa = 10.48TLTHH63 pKa = 6.23LRR65 pKa = 11.84CYY67 pKa = 10.27EE68 pKa = 3.8IAFLADD74 pKa = 3.18VASATDD80 pKa = 3.68WDD82 pKa = 3.7QCIAPGCPYY91 pKa = 10.32PNGWAADD98 pKa = 3.65SKK100 pKa = 10.86VRR102 pKa = 11.84VMWSDD107 pKa = 3.12NGAGGQFGTLDD118 pKa = 3.55SNGNFIPLDD127 pKa = 3.59PVVNAQQITHH137 pKa = 5.98YY138 pKa = 9.63RR139 pKa = 11.84APWDD143 pKa = 3.45RR144 pKa = 11.84PGIVHH149 pKa = 6.27ITLVVDD155 pKa = 5.46DD156 pKa = 4.89EE157 pKa = 4.41GLYY160 pKa = 10.81YY161 pKa = 10.41DD162 pKa = 5.7DD163 pKa = 5.64PPVSNNSDD171 pKa = 3.45NGNVTVWEE179 pKa = 4.5FWITPCPYY187 pKa = 9.62NWRR190 pKa = 11.84PIPNTTGPSFIAWVEE205 pKa = 3.97PEE207 pKa = 4.13VDD209 pKa = 3.47HH210 pKa = 7.1LGQPMSGIMTFNLISSTEE228 pKa = 3.84PGEE231 pKa = 4.2CLNSHH236 pKa = 6.87FSCPYY241 pKa = 11.15DD242 pKa = 3.37MDD244 pKa = 3.48IVQTEE249 pKa = 3.97EE250 pKa = 4.64GEE252 pKa = 4.4TFEE255 pKa = 5.31VNDD258 pKa = 4.05IHH260 pKa = 8.16DD261 pKa = 4.85HH262 pKa = 6.8DD263 pKa = 6.22ADD265 pKa = 3.87LQFPPEE271 pKa = 4.1LEE273 pKa = 3.92GMNVYY278 pKa = 10.28GSQEE282 pKa = 3.78EE283 pKa = 4.65DD284 pKa = 3.27EE285 pKa = 4.77EE286 pKa = 5.28GPYY289 pKa = 8.64THH291 pKa = 6.96NFTVAVTQQPTTVAEE306 pKa = 4.05VRR308 pKa = 11.84ILCFDD313 pKa = 3.53GGGYY317 pKa = 10.75GFLLAGIDD325 pKa = 3.85GFPPGSATARR335 pKa = 11.84RR336 pKa = 11.84TDD338 pKa = 3.13TGVIGSVEE346 pKa = 3.78IPYY349 pKa = 9.99DD350 pKa = 3.56ANGNFIADD358 pKa = 3.19VWEE361 pKa = 4.45ANMGIYY367 pKa = 9.77PADD370 pKa = 3.51SLGDD374 pKa = 3.61FDD376 pKa = 4.64NQPVGDD382 pKa = 4.03GTLGDD387 pKa = 3.74GLSVYY392 pKa = 10.33EE393 pKa = 4.4EE394 pKa = 4.01YY395 pKa = 10.58RR396 pKa = 11.84GLRR399 pKa = 11.84VQGIWTVFNPRR410 pKa = 11.84LKK412 pKa = 11.26DD413 pKa = 3.24MFVMNFGAGQEE424 pKa = 4.2DD425 pKa = 5.53PYY427 pKa = 11.76VPATIPNEE435 pKa = 4.47AITSQDD441 pKa = 3.13GFPGQGMPLLWLLNARR457 pKa = 11.84EE458 pKa = 4.07GLGYY462 pKa = 9.21PLRR465 pKa = 11.84VVNYY469 pKa = 8.78LNHH472 pKa = 6.15YY473 pKa = 8.8AHH475 pKa = 6.86IRR477 pKa = 11.84DD478 pKa = 3.61VYY480 pKa = 10.78AAIVVPGDD488 pKa = 3.12IEE490 pKa = 4.92GYY492 pKa = 10.27QDD494 pKa = 4.45ADD496 pKa = 3.71NDD498 pKa = 4.3GQPDD502 pKa = 3.8DD503 pKa = 4.13TNGDD507 pKa = 3.79GVIDD511 pKa = 4.71DD512 pKa = 4.52SDD514 pKa = 5.4LIVDD518 pKa = 3.55VRR520 pKa = 11.84AYY522 pKa = 10.44GFTYY526 pKa = 10.47GPIWDD531 pKa = 3.99NAGGPIAPPGDD542 pKa = 3.17WGYY545 pKa = 10.61IPQFWGPPVIEE556 pKa = 4.15VDD558 pKa = 2.92VDD560 pKa = 3.3YY561 pKa = 11.39LRR563 pKa = 11.84NEE565 pKa = 4.07RR566 pKa = 11.84LDD568 pKa = 3.93PNTNPNGLTEE578 pKa = 4.26LQVLTWVVGHH588 pKa = 6.03EE589 pKa = 4.32LGHH592 pKa = 5.27TVLWHH597 pKa = 6.34RR598 pKa = 11.84NDD600 pKa = 3.33LGGYY604 pKa = 6.92GHH606 pKa = 7.28HH607 pKa = 6.96NGFDD611 pKa = 3.42AAGNPITTYY620 pKa = 11.25DD621 pKa = 3.69CLIWQWMDD629 pKa = 2.56WRR631 pKa = 11.84ANPPTEE637 pKa = 4.06FCAVNPGDD645 pKa = 3.55QTRR648 pKa = 11.84WKK650 pKa = 10.24INPP653 pKa = 3.5
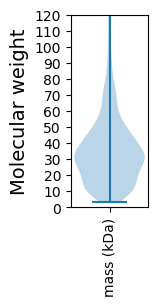
Molecular weight: 72.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5XEA0|A0A2H5XEA0_9BACT ECF RNA polymerase sigma-E factor OS=bacterium HR17 OX=2035412 GN=rpoE_4 PE=3 SV=1
MM1 pKa = 7.82WKK3 pKa = 10.64RR4 pKa = 11.84MMAWLLAGTMTLPVAVWAQQPSKK27 pKa = 11.09KK28 pKa = 8.65PTQPPAKK35 pKa = 9.12KK36 pKa = 9.95AQPAKK41 pKa = 9.56PAQPAKK47 pKa = 9.61PATPATPAKK56 pKa = 9.22PATPATPAKK65 pKa = 9.49PATPAQPATPAKK77 pKa = 10.08KK78 pKa = 10.13AATGTKK84 pKa = 9.11KK85 pKa = 10.14RR86 pKa = 11.84AAKK89 pKa = 9.66RR90 pKa = 11.84HH91 pKa = 4.11VRR93 pKa = 11.84KK94 pKa = 9.39HH95 pKa = 5.12RR96 pKa = 11.84KK97 pKa = 7.6VRR99 pKa = 11.84HH100 pKa = 5.49RR101 pKa = 11.84RR102 pKa = 11.84PAHH105 pKa = 6.03RR106 pKa = 11.84RR107 pKa = 11.84TTQKK111 pKa = 10.29VAAAKK116 pKa = 10.14KK117 pKa = 9.78
MM1 pKa = 7.82WKK3 pKa = 10.64RR4 pKa = 11.84MMAWLLAGTMTLPVAVWAQQPSKK27 pKa = 11.09KK28 pKa = 8.65PTQPPAKK35 pKa = 9.12KK36 pKa = 9.95AQPAKK41 pKa = 9.56PAQPAKK47 pKa = 9.61PATPATPAKK56 pKa = 9.22PATPATPAKK65 pKa = 9.49PATPAQPATPAKK77 pKa = 10.08KK78 pKa = 10.13AATGTKK84 pKa = 9.11KK85 pKa = 10.14RR86 pKa = 11.84AAKK89 pKa = 9.66RR90 pKa = 11.84HH91 pKa = 4.11VRR93 pKa = 11.84KK94 pKa = 9.39HH95 pKa = 5.12RR96 pKa = 11.84KK97 pKa = 7.6VRR99 pKa = 11.84HH100 pKa = 5.49RR101 pKa = 11.84RR102 pKa = 11.84PAHH105 pKa = 6.03RR106 pKa = 11.84RR107 pKa = 11.84TTQKK111 pKa = 10.29VAAAKK116 pKa = 10.14KK117 pKa = 9.78
Molecular weight: 12.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
939222 |
29 |
2022 |
337.7 |
37.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.825 ± 0.059 | 1.164 ± 0.016 |
4.859 ± 0.031 | 5.852 ± 0.051 |
3.851 ± 0.032 | 7.441 ± 0.042 |
2.299 ± 0.022 | 4.274 ± 0.031 |
3.172 ± 0.033 | 11.111 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.022 ± 0.019 | 2.39 ± 0.022 |
5.862 ± 0.04 | 4.134 ± 0.032 |
8.064 ± 0.042 | 4.035 ± 0.028 |
5.41 ± 0.034 | 8.782 ± 0.044 |
2.163 ± 0.027 | 2.291 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
