
Eutypa lata (strain UCR-EL1) (Grapevine dieback disease fungus) (Eutypa armeniacae)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Diatrypaceae; Eutypa; Eutypa lata
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
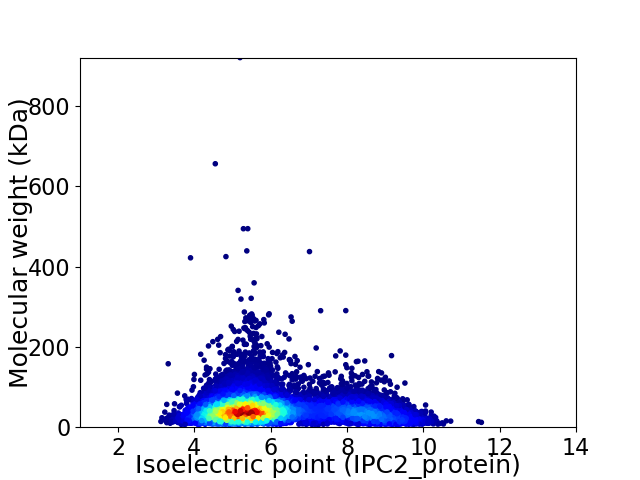
Virtual 2D-PAGE plot for 11684 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|M7TX45|M7TX45_EUTLA Dolichyl-phosphate-mannose--protein mannosyltransferase OS=Eutypa lata (strain UCR-EL1) OX=1287681 GN=UCREL1_1721 PE=3 SV=1
MM1 pKa = 6.98NAHH4 pKa = 7.47RR5 pKa = 11.84IAVDD9 pKa = 2.89IGTPKK14 pKa = 8.77QTVGLRR20 pKa = 11.84VDD22 pKa = 3.44VSSYY26 pKa = 6.96QTWAISGCGDD36 pKa = 3.62LFFNEE41 pKa = 4.78DD42 pKa = 3.68EE43 pKa = 4.32CDD45 pKa = 3.63KK46 pKa = 11.46AGTYY50 pKa = 10.39NDD52 pKa = 4.17SLSDD56 pKa = 3.18TSLYY60 pKa = 10.94VDD62 pKa = 3.53VPYY65 pKa = 10.9SEE67 pKa = 5.51SSVDD71 pKa = 3.57NDD73 pKa = 4.22DD74 pKa = 3.89GSSYY78 pKa = 11.43SLTYY82 pKa = 10.38YY83 pKa = 11.05ADD85 pKa = 4.02DD86 pKa = 3.73FTIQGGDD93 pKa = 3.24TLKK96 pKa = 11.09NITFGVFDD104 pKa = 5.16HH105 pKa = 7.15PNSDD109 pKa = 3.18QFGALGLGFGKK120 pKa = 10.59GVNSDD125 pKa = 3.03RR126 pKa = 11.84SNIVDD131 pKa = 3.61EE132 pKa = 5.25LVAQGFTQTKK142 pKa = 10.07AFSLSLGPYY151 pKa = 8.93GSEE154 pKa = 3.78EE155 pKa = 3.74ASLILGGVDD164 pKa = 3.05TKK166 pKa = 11.07KK167 pKa = 10.9FSGSLQRR174 pKa = 11.84MPIVDD179 pKa = 4.21GPEE182 pKa = 3.24FDD184 pKa = 4.39YY185 pKa = 11.46DD186 pKa = 4.46FNQTQYY192 pKa = 10.28WISIDD197 pKa = 3.44EE198 pKa = 4.36ATTNRR203 pKa = 11.84PDD205 pKa = 3.54ADD207 pKa = 3.5ATSFPSFKK215 pKa = 10.87AMQHH219 pKa = 5.44SDD221 pKa = 3.04IDD223 pKa = 4.42VSWLPWEE230 pKa = 4.15IAEE233 pKa = 4.9SVAGDD238 pKa = 3.92FGLAIAEE245 pKa = 5.1DD246 pKa = 3.74NYY248 pKa = 10.5EE249 pKa = 4.1WYY251 pKa = 9.71IVPCSAQNLTGSLDD265 pKa = 3.74LKK267 pKa = 11.09FGDD270 pKa = 4.0LALSIPYY277 pKa = 10.12GDD279 pKa = 5.32LVLQGDD285 pKa = 3.41IRR287 pKa = 11.84DD288 pKa = 3.9RR289 pKa = 11.84LVDD292 pKa = 3.52EE293 pKa = 5.77CYY295 pKa = 10.66LSVMPRR301 pKa = 11.84NATDD305 pKa = 3.17EE306 pKa = 4.37GPIYY310 pKa = 10.38WLGQNIMGHH319 pKa = 7.36LYY321 pKa = 9.88TVFDD325 pKa = 3.75QEE327 pKa = 5.24SRR329 pKa = 11.84AVWLAEE335 pKa = 3.54YY336 pKa = 9.86DD337 pKa = 3.56NCGTEE342 pKa = 4.15VVEE345 pKa = 4.13ITKK348 pKa = 10.53DD349 pKa = 3.17EE350 pKa = 4.03NAIAGINGQCGGPGIQAGGDD370 pKa = 3.61EE371 pKa = 4.61EE372 pKa = 4.28EE373 pKa = 5.1AGAGDD378 pKa = 3.74AEE380 pKa = 4.82GFGTRR385 pKa = 11.84INGPAVLLWLTSIFAGLLLGFF406 pKa = 5.07
MM1 pKa = 6.98NAHH4 pKa = 7.47RR5 pKa = 11.84IAVDD9 pKa = 2.89IGTPKK14 pKa = 8.77QTVGLRR20 pKa = 11.84VDD22 pKa = 3.44VSSYY26 pKa = 6.96QTWAISGCGDD36 pKa = 3.62LFFNEE41 pKa = 4.78DD42 pKa = 3.68EE43 pKa = 4.32CDD45 pKa = 3.63KK46 pKa = 11.46AGTYY50 pKa = 10.39NDD52 pKa = 4.17SLSDD56 pKa = 3.18TSLYY60 pKa = 10.94VDD62 pKa = 3.53VPYY65 pKa = 10.9SEE67 pKa = 5.51SSVDD71 pKa = 3.57NDD73 pKa = 4.22DD74 pKa = 3.89GSSYY78 pKa = 11.43SLTYY82 pKa = 10.38YY83 pKa = 11.05ADD85 pKa = 4.02DD86 pKa = 3.73FTIQGGDD93 pKa = 3.24TLKK96 pKa = 11.09NITFGVFDD104 pKa = 5.16HH105 pKa = 7.15PNSDD109 pKa = 3.18QFGALGLGFGKK120 pKa = 10.59GVNSDD125 pKa = 3.03RR126 pKa = 11.84SNIVDD131 pKa = 3.61EE132 pKa = 5.25LVAQGFTQTKK142 pKa = 10.07AFSLSLGPYY151 pKa = 8.93GSEE154 pKa = 3.78EE155 pKa = 3.74ASLILGGVDD164 pKa = 3.05TKK166 pKa = 11.07KK167 pKa = 10.9FSGSLQRR174 pKa = 11.84MPIVDD179 pKa = 4.21GPEE182 pKa = 3.24FDD184 pKa = 4.39YY185 pKa = 11.46DD186 pKa = 4.46FNQTQYY192 pKa = 10.28WISIDD197 pKa = 3.44EE198 pKa = 4.36ATTNRR203 pKa = 11.84PDD205 pKa = 3.54ADD207 pKa = 3.5ATSFPSFKK215 pKa = 10.87AMQHH219 pKa = 5.44SDD221 pKa = 3.04IDD223 pKa = 4.42VSWLPWEE230 pKa = 4.15IAEE233 pKa = 4.9SVAGDD238 pKa = 3.92FGLAIAEE245 pKa = 5.1DD246 pKa = 3.74NYY248 pKa = 10.5EE249 pKa = 4.1WYY251 pKa = 9.71IVPCSAQNLTGSLDD265 pKa = 3.74LKK267 pKa = 11.09FGDD270 pKa = 4.0LALSIPYY277 pKa = 10.12GDD279 pKa = 5.32LVLQGDD285 pKa = 3.41IRR287 pKa = 11.84DD288 pKa = 3.9RR289 pKa = 11.84LVDD292 pKa = 3.52EE293 pKa = 5.77CYY295 pKa = 10.66LSVMPRR301 pKa = 11.84NATDD305 pKa = 3.17EE306 pKa = 4.37GPIYY310 pKa = 10.38WLGQNIMGHH319 pKa = 7.36LYY321 pKa = 9.88TVFDD325 pKa = 3.75QEE327 pKa = 5.24SRR329 pKa = 11.84AVWLAEE335 pKa = 3.54YY336 pKa = 9.86DD337 pKa = 3.56NCGTEE342 pKa = 4.15VVEE345 pKa = 4.13ITKK348 pKa = 10.53DD349 pKa = 3.17EE350 pKa = 4.03NAIAGINGQCGGPGIQAGGDD370 pKa = 3.61EE371 pKa = 4.61EE372 pKa = 4.28EE373 pKa = 5.1AGAGDD378 pKa = 3.74AEE380 pKa = 4.82GFGTRR385 pKa = 11.84INGPAVLLWLTSIFAGLLLGFF406 pKa = 5.07
Molecular weight: 43.98 kDa
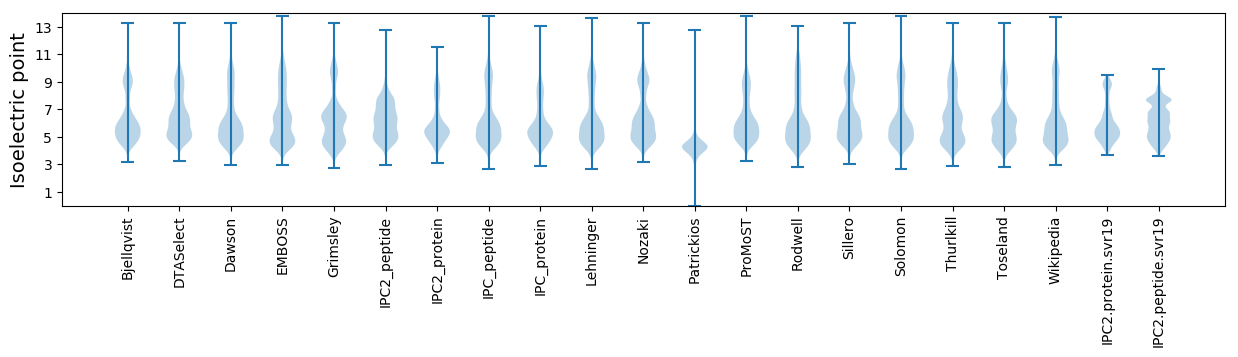
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7SVI8|M7SVI8_EUTLA Putative atpase family aaa domain-containing protein 1-a protein OS=Eutypa lata (strain UCR-EL1) OX=1287681 GN=UCREL1_4440 PE=4 SV=1
MM1 pKa = 7.94PKK3 pKa = 9.44QQRR6 pKa = 11.84TLKK9 pKa = 9.11QQRR12 pKa = 11.84TLKK15 pKa = 10.15QQLTPKK21 pKa = 9.49QLKK24 pKa = 9.96RR25 pKa = 11.84RR26 pKa = 11.84IKK28 pKa = 9.55QQPTSKK34 pKa = 10.23QLKK37 pKa = 8.78RR38 pKa = 11.84QLKK41 pKa = 7.34RR42 pKa = 11.84QRR44 pKa = 11.84MPKK47 pKa = 9.25QLKK50 pKa = 7.12QQRR53 pKa = 11.84TPKK56 pKa = 9.94QQLTPKK62 pKa = 9.7QLKK65 pKa = 9.84RR66 pKa = 11.84QLKK69 pKa = 8.61QQLTPKK75 pKa = 9.73QLKK78 pKa = 9.76RR79 pKa = 11.84QLKK82 pKa = 7.34RR83 pKa = 11.84QRR85 pKa = 11.84MPKK88 pKa = 9.27QLKK91 pKa = 8.24QQQTLKK97 pKa = 10.49RR98 pKa = 11.84QRR100 pKa = 11.84TLKK103 pKa = 10.37RR104 pKa = 11.84QQTPKK109 pKa = 10.27QPKK112 pKa = 8.59
MM1 pKa = 7.94PKK3 pKa = 9.44QQRR6 pKa = 11.84TLKK9 pKa = 9.11QQRR12 pKa = 11.84TLKK15 pKa = 10.15QQLTPKK21 pKa = 9.49QLKK24 pKa = 9.96RR25 pKa = 11.84RR26 pKa = 11.84IKK28 pKa = 9.55QQPTSKK34 pKa = 10.23QLKK37 pKa = 8.78RR38 pKa = 11.84QLKK41 pKa = 7.34RR42 pKa = 11.84QRR44 pKa = 11.84MPKK47 pKa = 9.25QLKK50 pKa = 7.12QQRR53 pKa = 11.84TPKK56 pKa = 9.94QQLTPKK62 pKa = 9.7QLKK65 pKa = 9.84RR66 pKa = 11.84QLKK69 pKa = 8.61QQLTPKK75 pKa = 9.73QLKK78 pKa = 9.76RR79 pKa = 11.84QLKK82 pKa = 7.34RR83 pKa = 11.84QRR85 pKa = 11.84MPKK88 pKa = 9.27QLKK91 pKa = 8.24QQQTLKK97 pKa = 10.49RR98 pKa = 11.84QRR100 pKa = 11.84TLKK103 pKa = 10.37RR104 pKa = 11.84QQTPKK109 pKa = 10.27QPKK112 pKa = 8.59
Molecular weight: 13.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5038052 |
66 |
8334 |
431.2 |
47.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.176 ± 0.022 | 1.13 ± 0.008 |
6.127 ± 0.016 | 6.418 ± 0.027 |
3.746 ± 0.014 | 7.6 ± 0.026 |
2.243 ± 0.01 | 4.806 ± 0.017 |
4.775 ± 0.021 | 8.708 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.152 ± 0.009 | 3.669 ± 0.013 |
5.578 ± 0.022 | 3.795 ± 0.016 |
5.847 ± 0.023 | 7.46 ± 0.022 |
5.946 ± 0.018 | 6.472 ± 0.017 |
1.538 ± 0.009 | 2.814 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
