
Epulopiscium sp. Nele67-Bin004
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Epulopiscium; unclassified Epulopiscium
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
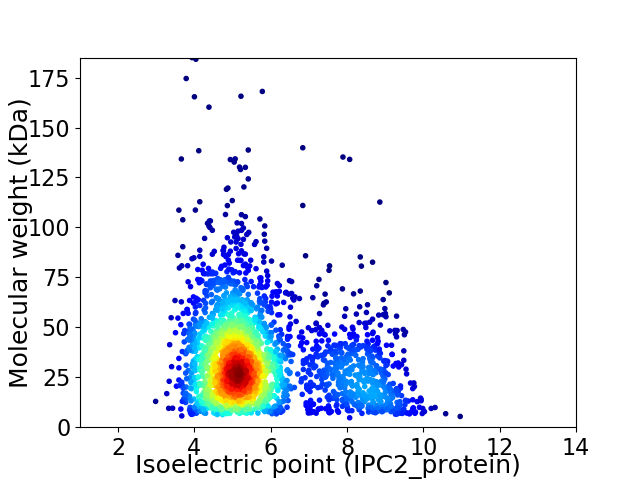
Virtual 2D-PAGE plot for 2474 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
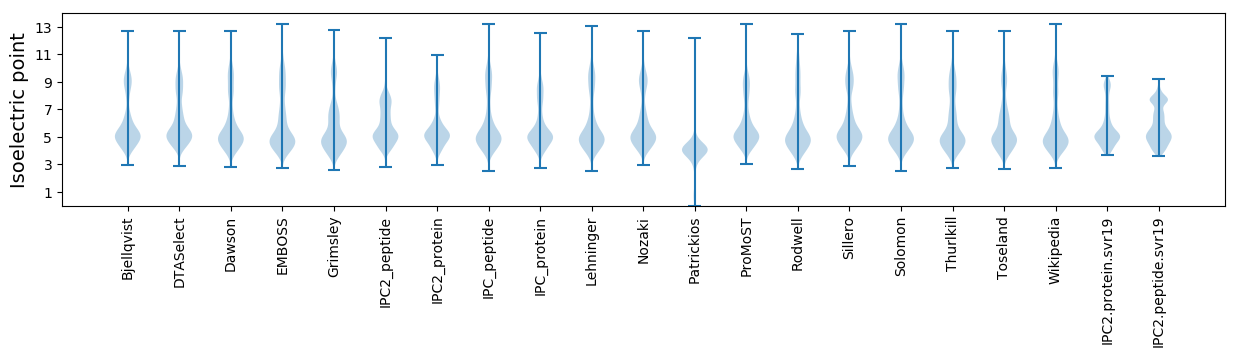
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S9CNY1|A0A1S9CNY1_9FIRM Fido domain-containing protein OS=Epulopiscium sp. Nele67-Bin004 OX=1764962 GN=ATN35_04455 PE=4 SV=1
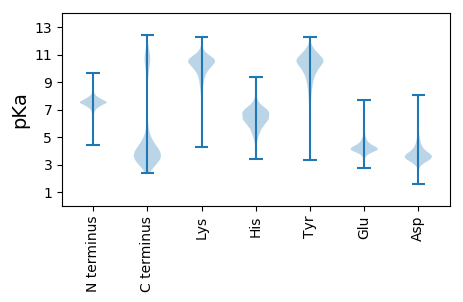
MM1 pKa = 7.84KK2 pKa = 10.51LGTKK6 pKa = 9.05LRR8 pKa = 11.84KK9 pKa = 9.36RR10 pKa = 11.84LAFLLASSIIFASTSSINSANYY32 pKa = 8.38TRR34 pKa = 11.84QQTNSTIIDD43 pKa = 3.33VGYY46 pKa = 10.09IQINEE51 pKa = 3.84NEE53 pKa = 4.69LYY55 pKa = 10.5YY56 pKa = 11.01FPDD59 pKa = 3.32LTVNIPNNNKK69 pKa = 9.06VFGLNLFYY77 pKa = 11.13DD78 pKa = 4.59DD79 pKa = 4.52ANKK82 pKa = 10.95DD83 pKa = 3.6GVDD86 pKa = 4.53GILSLVDD93 pKa = 3.6PQIEE97 pKa = 4.28GLTYY101 pKa = 10.81TKK103 pKa = 8.11MTNLDD108 pKa = 3.76GEE110 pKa = 4.51QGSVLYY116 pKa = 10.43AFSGGIDD123 pKa = 3.25AQTAQEE129 pKa = 4.36FLRR132 pKa = 11.84ALQFDD137 pKa = 3.77ITGNTPLNIIVDD149 pKa = 3.89QNEE152 pKa = 4.11VVLPEE157 pKa = 4.48SEE159 pKa = 3.96NEE161 pKa = 3.95IYY163 pKa = 8.21FTKK166 pKa = 10.6YY167 pKa = 9.98IPEE170 pKa = 4.99DD171 pKa = 3.63DD172 pKa = 4.47PEE174 pKa = 4.13NVHH177 pKa = 6.35YY178 pKa = 10.51YY179 pKa = 10.27FYY181 pKa = 11.52VNEE184 pKa = 4.03LVDD187 pKa = 3.14WDD189 pKa = 4.09TAYY192 pKa = 10.43EE193 pKa = 3.93LAKK196 pKa = 10.13SYY198 pKa = 10.86RR199 pKa = 11.84YY200 pKa = 9.6MGMKK204 pKa = 10.22GYY206 pKa = 10.01LATITTPGEE215 pKa = 4.16DD216 pKa = 3.27QLLTSISKK224 pKa = 10.66LGGWTSGIRR233 pKa = 11.84IDD235 pKa = 3.47PTYY238 pKa = 11.11LFGEE242 pKa = 4.39NGVDD246 pKa = 4.12SDD248 pKa = 4.62TISANLTAKK257 pKa = 9.89TGIFNSGLGHH267 pKa = 6.93KK268 pKa = 10.01SNSGHH273 pKa = 5.34VQLIDD278 pKa = 3.27RR279 pKa = 11.84FYY281 pKa = 9.75WAAGPEE287 pKa = 4.33AIAQKK292 pKa = 11.1DD293 pKa = 4.06DD294 pKa = 3.46IGDD297 pKa = 3.49MTKK300 pKa = 11.01NNWGFYY306 pKa = 10.65NYY308 pKa = 10.46DD309 pKa = 3.46GTNGTTSSAIRR320 pKa = 11.84WDD322 pKa = 3.59NGYY325 pKa = 10.44EE326 pKa = 3.89PYY328 pKa = 11.0QNFTKK333 pKa = 10.78ANGEE337 pKa = 4.05NEE339 pKa = 4.2PNDD342 pKa = 3.87GGTVATKK349 pKa = 10.28EE350 pKa = 4.22YY351 pKa = 9.9IVQVNWGGTNVFAWNDD367 pKa = 3.46LAAKK371 pKa = 8.27EE372 pKa = 3.91LWIRR376 pKa = 11.84GYY378 pKa = 9.99FVEE381 pKa = 4.24MSPYY385 pKa = 10.05EE386 pKa = 4.27GGVVPGYY393 pKa = 9.88EE394 pKa = 4.09DD395 pKa = 5.21NIIEE399 pKa = 4.24TNINIIMEE407 pKa = 4.76DD408 pKa = 3.96LPVYY412 pKa = 10.72DD413 pKa = 5.27PDD415 pKa = 3.8YY416 pKa = 11.17QPTEE420 pKa = 4.29FPDD423 pKa = 3.65EE424 pKa = 4.49LPTPEE429 pKa = 4.92LDD431 pKa = 4.15GMVDD435 pKa = 4.34DD436 pKa = 4.99VFPDD440 pKa = 3.65EE441 pKa = 5.42LPTPEE446 pKa = 4.92LDD448 pKa = 4.15GMVDD452 pKa = 3.63DD453 pKa = 4.89VFPNEE458 pKa = 4.43LPTPEE463 pKa = 4.77LDD465 pKa = 3.46EE466 pKa = 5.71AGEE469 pKa = 4.14VLFPDD474 pKa = 4.23VLPTPDD480 pKa = 5.36LDD482 pKa = 4.12GLDD485 pKa = 4.78DD486 pKa = 3.94VFSDD490 pKa = 3.81VLPMPEE496 pKa = 4.62LDD498 pKa = 3.55GTIDD502 pKa = 4.36DD503 pKa = 4.53VFPDD507 pKa = 4.08VLPMPEE513 pKa = 4.58LDD515 pKa = 3.55GTIDD519 pKa = 4.36DD520 pKa = 4.53VFPDD524 pKa = 4.08VLPMPEE530 pKa = 5.2LDD532 pKa = 4.15GLDD535 pKa = 3.57DD536 pKa = 3.8VLYY539 pKa = 10.28PDD541 pKa = 4.55MLPTPEE547 pKa = 5.58LDD549 pKa = 3.76GLDD552 pKa = 3.6DD553 pKa = 3.7VLYY556 pKa = 10.19PDD558 pKa = 4.51VLPTPEE564 pKa = 5.48LDD566 pKa = 3.82GLDD569 pKa = 3.6DD570 pKa = 3.7VLYY573 pKa = 10.19PDD575 pKa = 4.51VLPTPEE581 pKa = 5.48LDD583 pKa = 3.94GLDD586 pKa = 4.04DD587 pKa = 4.06VLFPDD592 pKa = 4.85EE593 pKa = 5.42LPMPEE598 pKa = 4.61LFPYY602 pKa = 10.32VEE604 pKa = 5.18PDD606 pKa = 3.24NDD608 pKa = 3.79DD609 pKa = 5.46DD610 pKa = 5.54KK611 pKa = 12.13AEE613 pKa = 5.1DD614 pKa = 4.87DD615 pKa = 4.76NSSDD619 pKa = 4.08DD620 pKa = 3.83NTAEE624 pKa = 4.23TEE626 pKa = 4.0NNTNYY631 pKa = 10.27EE632 pKa = 4.18IKK634 pKa = 10.41DD635 pKa = 3.45KK636 pKa = 11.48NDD638 pKa = 3.54IIEE641 pKa = 3.96QDD643 pKa = 3.36EE644 pKa = 4.64PIILEE649 pKa = 4.04LEE651 pKa = 3.96ARR653 pKa = 11.84MSRR656 pKa = 11.84MLSGFEE662 pKa = 3.91IDD664 pKa = 6.0IISDD668 pKa = 3.18WDD670 pKa = 3.66FEE672 pKa = 4.56IDD674 pKa = 4.2IIEE677 pKa = 4.68TILNAKK683 pKa = 9.49VDD685 pKa = 4.32LEE687 pKa = 4.92LIDD690 pKa = 5.51AIIDD694 pKa = 3.47AGIEE698 pKa = 4.01PEE700 pKa = 3.97IVEE703 pKa = 4.2QLVCGHH709 pKa = 7.01IDD711 pKa = 3.42AEE713 pKa = 4.78QFKK716 pKa = 9.51QLCNDD721 pKa = 3.32YY722 pKa = 9.75EE723 pKa = 4.27INIIYY728 pKa = 9.33EE729 pKa = 3.88EE730 pKa = 4.74PYY732 pKa = 9.89IFGYY736 pKa = 9.84PDD738 pKa = 3.29YY739 pKa = 11.52TFMTDD744 pKa = 3.06NNLTRR749 pKa = 11.84AEE751 pKa = 4.11VVAIVSRR758 pKa = 11.84LHH760 pKa = 5.92KK761 pKa = 10.88VEE763 pKa = 3.87MDD765 pKa = 3.22EE766 pKa = 4.26TATYY770 pKa = 9.33TSSFNDD776 pKa = 3.32VYY778 pKa = 9.53PTNWHH783 pKa = 6.29ANSIGFVEE791 pKa = 4.25QQGILNGYY799 pKa = 9.17KK800 pKa = 10.43DD801 pKa = 4.39GSFKK805 pKa = 11.04PNVAITRR812 pKa = 11.84AEE814 pKa = 4.63FIALTAPLGVATEE827 pKa = 4.21PKK829 pKa = 9.97VDD831 pKa = 3.88YY832 pKa = 10.83FPFDD836 pKa = 4.61DD837 pKa = 4.13VSQSHH842 pKa = 5.79WAYY845 pKa = 9.96EE846 pKa = 4.27SLHH849 pKa = 5.84IAYY852 pKa = 9.85SQGWVAGYY860 pKa = 10.7EE861 pKa = 4.07DD862 pKa = 3.82GKK864 pKa = 10.96YY865 pKa = 9.86RR866 pKa = 11.84PDD868 pKa = 3.22EE869 pKa = 4.83AITRR873 pKa = 11.84DD874 pKa = 3.39EE875 pKa = 4.05AVAIINRR882 pKa = 11.84IIDD885 pKa = 3.57RR886 pKa = 11.84RR887 pKa = 11.84PNPMFFDD894 pKa = 4.14VIFDD898 pKa = 4.39DD899 pKa = 3.65MHH901 pKa = 6.17EE902 pKa = 4.31SHH904 pKa = 6.84WAYY907 pKa = 10.44EE908 pKa = 3.9DD909 pKa = 4.17VILSSNYY916 pKa = 10.07YY917 pKa = 9.75IRR919 pKa = 11.84LDD921 pKa = 3.41KK922 pKa = 11.29
MM1 pKa = 7.84KK2 pKa = 10.51LGTKK6 pKa = 9.05LRR8 pKa = 11.84KK9 pKa = 9.36RR10 pKa = 11.84LAFLLASSIIFASTSSINSANYY32 pKa = 8.38TRR34 pKa = 11.84QQTNSTIIDD43 pKa = 3.33VGYY46 pKa = 10.09IQINEE51 pKa = 3.84NEE53 pKa = 4.69LYY55 pKa = 10.5YY56 pKa = 11.01FPDD59 pKa = 3.32LTVNIPNNNKK69 pKa = 9.06VFGLNLFYY77 pKa = 11.13DD78 pKa = 4.59DD79 pKa = 4.52ANKK82 pKa = 10.95DD83 pKa = 3.6GVDD86 pKa = 4.53GILSLVDD93 pKa = 3.6PQIEE97 pKa = 4.28GLTYY101 pKa = 10.81TKK103 pKa = 8.11MTNLDD108 pKa = 3.76GEE110 pKa = 4.51QGSVLYY116 pKa = 10.43AFSGGIDD123 pKa = 3.25AQTAQEE129 pKa = 4.36FLRR132 pKa = 11.84ALQFDD137 pKa = 3.77ITGNTPLNIIVDD149 pKa = 3.89QNEE152 pKa = 4.11VVLPEE157 pKa = 4.48SEE159 pKa = 3.96NEE161 pKa = 3.95IYY163 pKa = 8.21FTKK166 pKa = 10.6YY167 pKa = 9.98IPEE170 pKa = 4.99DD171 pKa = 3.63DD172 pKa = 4.47PEE174 pKa = 4.13NVHH177 pKa = 6.35YY178 pKa = 10.51YY179 pKa = 10.27FYY181 pKa = 11.52VNEE184 pKa = 4.03LVDD187 pKa = 3.14WDD189 pKa = 4.09TAYY192 pKa = 10.43EE193 pKa = 3.93LAKK196 pKa = 10.13SYY198 pKa = 10.86RR199 pKa = 11.84YY200 pKa = 9.6MGMKK204 pKa = 10.22GYY206 pKa = 10.01LATITTPGEE215 pKa = 4.16DD216 pKa = 3.27QLLTSISKK224 pKa = 10.66LGGWTSGIRR233 pKa = 11.84IDD235 pKa = 3.47PTYY238 pKa = 11.11LFGEE242 pKa = 4.39NGVDD246 pKa = 4.12SDD248 pKa = 4.62TISANLTAKK257 pKa = 9.89TGIFNSGLGHH267 pKa = 6.93KK268 pKa = 10.01SNSGHH273 pKa = 5.34VQLIDD278 pKa = 3.27RR279 pKa = 11.84FYY281 pKa = 9.75WAAGPEE287 pKa = 4.33AIAQKK292 pKa = 11.1DD293 pKa = 4.06DD294 pKa = 3.46IGDD297 pKa = 3.49MTKK300 pKa = 11.01NNWGFYY306 pKa = 10.65NYY308 pKa = 10.46DD309 pKa = 3.46GTNGTTSSAIRR320 pKa = 11.84WDD322 pKa = 3.59NGYY325 pKa = 10.44EE326 pKa = 3.89PYY328 pKa = 11.0QNFTKK333 pKa = 10.78ANGEE337 pKa = 4.05NEE339 pKa = 4.2PNDD342 pKa = 3.87GGTVATKK349 pKa = 10.28EE350 pKa = 4.22YY351 pKa = 9.9IVQVNWGGTNVFAWNDD367 pKa = 3.46LAAKK371 pKa = 8.27EE372 pKa = 3.91LWIRR376 pKa = 11.84GYY378 pKa = 9.99FVEE381 pKa = 4.24MSPYY385 pKa = 10.05EE386 pKa = 4.27GGVVPGYY393 pKa = 9.88EE394 pKa = 4.09DD395 pKa = 5.21NIIEE399 pKa = 4.24TNINIIMEE407 pKa = 4.76DD408 pKa = 3.96LPVYY412 pKa = 10.72DD413 pKa = 5.27PDD415 pKa = 3.8YY416 pKa = 11.17QPTEE420 pKa = 4.29FPDD423 pKa = 3.65EE424 pKa = 4.49LPTPEE429 pKa = 4.92LDD431 pKa = 4.15GMVDD435 pKa = 4.34DD436 pKa = 4.99VFPDD440 pKa = 3.65EE441 pKa = 5.42LPTPEE446 pKa = 4.92LDD448 pKa = 4.15GMVDD452 pKa = 3.63DD453 pKa = 4.89VFPNEE458 pKa = 4.43LPTPEE463 pKa = 4.77LDD465 pKa = 3.46EE466 pKa = 5.71AGEE469 pKa = 4.14VLFPDD474 pKa = 4.23VLPTPDD480 pKa = 5.36LDD482 pKa = 4.12GLDD485 pKa = 4.78DD486 pKa = 3.94VFSDD490 pKa = 3.81VLPMPEE496 pKa = 4.62LDD498 pKa = 3.55GTIDD502 pKa = 4.36DD503 pKa = 4.53VFPDD507 pKa = 4.08VLPMPEE513 pKa = 4.58LDD515 pKa = 3.55GTIDD519 pKa = 4.36DD520 pKa = 4.53VFPDD524 pKa = 4.08VLPMPEE530 pKa = 5.2LDD532 pKa = 4.15GLDD535 pKa = 3.57DD536 pKa = 3.8VLYY539 pKa = 10.28PDD541 pKa = 4.55MLPTPEE547 pKa = 5.58LDD549 pKa = 3.76GLDD552 pKa = 3.6DD553 pKa = 3.7VLYY556 pKa = 10.19PDD558 pKa = 4.51VLPTPEE564 pKa = 5.48LDD566 pKa = 3.82GLDD569 pKa = 3.6DD570 pKa = 3.7VLYY573 pKa = 10.19PDD575 pKa = 4.51VLPTPEE581 pKa = 5.48LDD583 pKa = 3.94GLDD586 pKa = 4.04DD587 pKa = 4.06VLFPDD592 pKa = 4.85EE593 pKa = 5.42LPMPEE598 pKa = 4.61LFPYY602 pKa = 10.32VEE604 pKa = 5.18PDD606 pKa = 3.24NDD608 pKa = 3.79DD609 pKa = 5.46DD610 pKa = 5.54KK611 pKa = 12.13AEE613 pKa = 5.1DD614 pKa = 4.87DD615 pKa = 4.76NSSDD619 pKa = 4.08DD620 pKa = 3.83NTAEE624 pKa = 4.23TEE626 pKa = 4.0NNTNYY631 pKa = 10.27EE632 pKa = 4.18IKK634 pKa = 10.41DD635 pKa = 3.45KK636 pKa = 11.48NDD638 pKa = 3.54IIEE641 pKa = 3.96QDD643 pKa = 3.36EE644 pKa = 4.64PIILEE649 pKa = 4.04LEE651 pKa = 3.96ARR653 pKa = 11.84MSRR656 pKa = 11.84MLSGFEE662 pKa = 3.91IDD664 pKa = 6.0IISDD668 pKa = 3.18WDD670 pKa = 3.66FEE672 pKa = 4.56IDD674 pKa = 4.2IIEE677 pKa = 4.68TILNAKK683 pKa = 9.49VDD685 pKa = 4.32LEE687 pKa = 4.92LIDD690 pKa = 5.51AIIDD694 pKa = 3.47AGIEE698 pKa = 4.01PEE700 pKa = 3.97IVEE703 pKa = 4.2QLVCGHH709 pKa = 7.01IDD711 pKa = 3.42AEE713 pKa = 4.78QFKK716 pKa = 9.51QLCNDD721 pKa = 3.32YY722 pKa = 9.75EE723 pKa = 4.27INIIYY728 pKa = 9.33EE729 pKa = 3.88EE730 pKa = 4.74PYY732 pKa = 9.89IFGYY736 pKa = 9.84PDD738 pKa = 3.29YY739 pKa = 11.52TFMTDD744 pKa = 3.06NNLTRR749 pKa = 11.84AEE751 pKa = 4.11VVAIVSRR758 pKa = 11.84LHH760 pKa = 5.92KK761 pKa = 10.88VEE763 pKa = 3.87MDD765 pKa = 3.22EE766 pKa = 4.26TATYY770 pKa = 9.33TSSFNDD776 pKa = 3.32VYY778 pKa = 9.53PTNWHH783 pKa = 6.29ANSIGFVEE791 pKa = 4.25QQGILNGYY799 pKa = 9.17KK800 pKa = 10.43DD801 pKa = 4.39GSFKK805 pKa = 11.04PNVAITRR812 pKa = 11.84AEE814 pKa = 4.63FIALTAPLGVATEE827 pKa = 4.21PKK829 pKa = 9.97VDD831 pKa = 3.88YY832 pKa = 10.83FPFDD836 pKa = 4.61DD837 pKa = 4.13VSQSHH842 pKa = 5.79WAYY845 pKa = 9.96EE846 pKa = 4.27SLHH849 pKa = 5.84IAYY852 pKa = 9.85SQGWVAGYY860 pKa = 10.7EE861 pKa = 4.07DD862 pKa = 3.82GKK864 pKa = 10.96YY865 pKa = 9.86RR866 pKa = 11.84PDD868 pKa = 3.22EE869 pKa = 4.83AITRR873 pKa = 11.84DD874 pKa = 3.39EE875 pKa = 4.05AVAIINRR882 pKa = 11.84IIDD885 pKa = 3.57RR886 pKa = 11.84RR887 pKa = 11.84PNPMFFDD894 pKa = 4.14VIFDD898 pKa = 4.39DD899 pKa = 3.65MHH901 pKa = 6.17EE902 pKa = 4.31SHH904 pKa = 6.84WAYY907 pKa = 10.44EE908 pKa = 3.9DD909 pKa = 4.17VILSSNYY916 pKa = 10.07YY917 pKa = 9.75IRR919 pKa = 11.84LDD921 pKa = 3.41KK922 pKa = 11.29
Molecular weight: 103.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S9CQV6|A0A1S9CQV6_9FIRM Uncharacterized protein OS=Epulopiscium sp. Nele67-Bin004 OX=1764962 GN=ATN35_00150 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.49KK9 pKa = 7.5RR10 pKa = 11.84QRR12 pKa = 11.84SRR14 pKa = 11.84EE15 pKa = 3.65HH16 pKa = 6.35GFRR19 pKa = 11.84KK20 pKa = 9.98RR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 10.42NGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 9.04KK41 pKa = 9.67LTAA44 pKa = 4.2
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.49KK9 pKa = 7.5RR10 pKa = 11.84QRR12 pKa = 11.84SRR14 pKa = 11.84EE15 pKa = 3.65HH16 pKa = 6.35GFRR19 pKa = 11.84KK20 pKa = 9.98RR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 10.42NGRR28 pKa = 11.84AVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 9.04KK41 pKa = 9.67LTAA44 pKa = 4.2
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
779308 |
44 |
1661 |
315.0 |
35.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.151 ± 0.051 | 1.08 ± 0.021 |
6.02 ± 0.038 | 7.248 ± 0.052 |
4.129 ± 0.034 | 6.052 ± 0.041 |
1.634 ± 0.024 | 9.138 ± 0.047 |
6.938 ± 0.055 | 9.022 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.727 ± 0.023 | 5.617 ± 0.038 |
2.987 ± 0.037 | 3.619 ± 0.029 |
3.217 ± 0.031 | 6.174 ± 0.043 |
6.24 ± 0.051 | 6.943 ± 0.044 |
0.732 ± 0.014 | 4.333 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
