
Wenling triplecross lizardfish paramyxovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Metaparamyxovirinae; Synodonvirus; Synodus synodonvirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
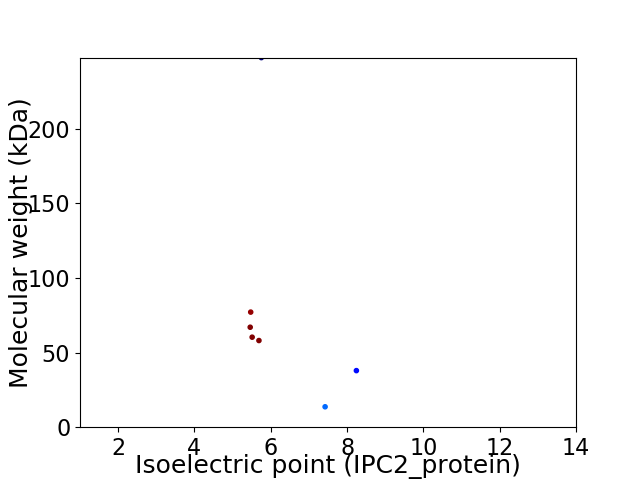
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
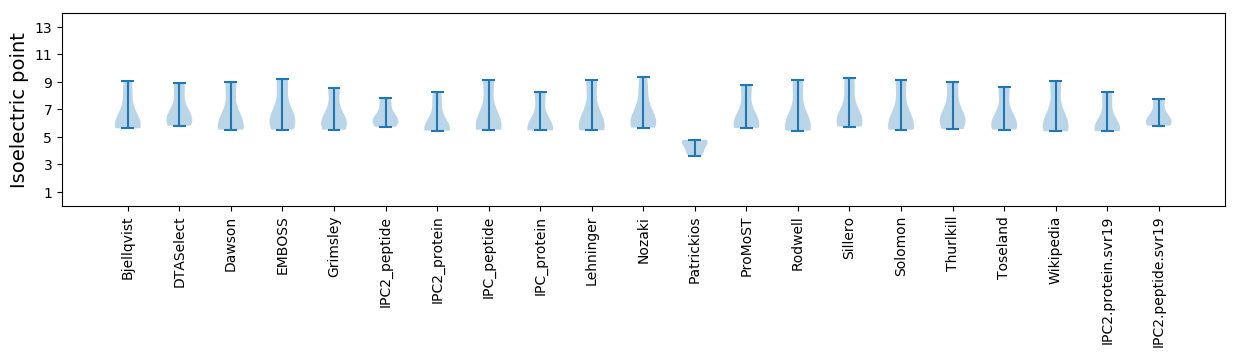
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GNG5|A0A2P1GNG5_9MONO RNA-directed RNA polymerase L OS=Wenling triplecross lizardfish paramyxovirus OX=2116451 PE=3 SV=1
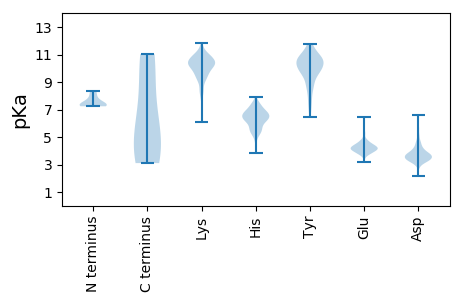
MM1 pKa = 7.32KK2 pKa = 10.29RR3 pKa = 11.84VLDD6 pKa = 3.71WVHH9 pKa = 6.34QKK11 pKa = 7.91MARR14 pKa = 11.84KK15 pKa = 9.49GLDD18 pKa = 3.89PINPTIKK25 pKa = 10.09VGRR28 pKa = 11.84DD29 pKa = 3.2GTEE32 pKa = 3.43VDD34 pKa = 3.44QGLAIYY40 pKa = 9.6IARR43 pKa = 11.84VTVSILAIILLTVLLVTPTVTGLFQVFDD71 pKa = 4.07SSASEE76 pKa = 3.87QSHH79 pKa = 7.37PIMDD83 pKa = 5.11LLNDD87 pKa = 3.66EE88 pKa = 4.7VLPPLRR94 pKa = 11.84NTHH97 pKa = 6.59ALLEE101 pKa = 4.5EE102 pKa = 4.36DD103 pKa = 3.25TAPRR107 pKa = 11.84VRR109 pKa = 11.84SIQTAVEE116 pKa = 3.69LRR118 pKa = 11.84IPGDD122 pKa = 3.61LKK124 pKa = 11.22SATQQILHH132 pKa = 7.31AIKK135 pKa = 9.08DD136 pKa = 3.8TCVGGDD142 pKa = 3.46GGGTGPAPSCPPEE155 pKa = 4.43GVLPQHH161 pKa = 6.72HH162 pKa = 6.63SAFKK166 pKa = 10.47PIDD169 pKa = 4.07PDD171 pKa = 3.78SFTPCATGKK180 pKa = 8.35PTFSGSPAASSSIGSFPWISSGTQDD205 pKa = 3.24TPKK208 pKa = 10.44ISMTMSSSYY217 pKa = 11.01LGFYY221 pKa = 9.57ATDD224 pKa = 3.54TYY226 pKa = 10.71ITTGQNKK233 pKa = 9.61RR234 pKa = 11.84SVSVIGLGRR243 pKa = 11.84LTVTEE248 pKa = 4.85DD249 pKa = 3.27GRR251 pKa = 11.84SSAVVTEE258 pKa = 4.64AEE260 pKa = 4.78PIHH263 pKa = 6.38HH264 pKa = 6.9SSLIGYY270 pKa = 8.77QSTGTGITGTVSFFRR285 pKa = 11.84TTGSEE290 pKa = 4.08DD291 pKa = 3.28TADD294 pKa = 3.82AQDD297 pKa = 3.37VGIRR301 pKa = 11.84SHH303 pKa = 6.24TLLVMRR309 pKa = 11.84RR310 pKa = 11.84NRR312 pKa = 11.84KK313 pKa = 9.08LDD315 pKa = 3.31TYY317 pKa = 10.16VYY319 pKa = 10.32EE320 pKa = 4.54GGEE323 pKa = 3.97IQGGDD328 pKa = 4.39MYY330 pKa = 11.37MMASPLSGQGTMVGDD345 pKa = 3.72EE346 pKa = 4.31MVLLGYY352 pKa = 10.07AVPVTNYY359 pKa = 10.0AGPVWCPNTFCNTQTMEE376 pKa = 3.91ACKK379 pKa = 10.14NASGTIRR386 pKa = 11.84FPGSSTGLAVVMVRR400 pKa = 11.84YY401 pKa = 8.7MIQDD405 pKa = 3.66LDD407 pKa = 3.81KK408 pKa = 11.61DD409 pKa = 3.88GFKK412 pKa = 10.83PKK414 pKa = 9.14ITIHH418 pKa = 5.28MLNRR422 pKa = 11.84EE423 pKa = 4.02QWPIPGPYY431 pKa = 9.71HH432 pKa = 5.82LTSGTKK438 pKa = 9.73GLFISVYY445 pKa = 7.86SWGWYY450 pKa = 6.75GHH452 pKa = 6.1PVSGEE457 pKa = 3.8INRR460 pKa = 11.84ATWDD464 pKa = 3.3MRR466 pKa = 11.84SIRR469 pKa = 11.84LVYY472 pKa = 10.54ALLWPDD478 pKa = 3.48LPEE481 pKa = 4.45TEE483 pKa = 4.5GKK485 pKa = 10.77CYY487 pKa = 10.5TNLANPCPRR496 pKa = 11.84ACTIGSTRR504 pKa = 11.84IGSGIPLTFTGQLSAWLTISADD526 pKa = 3.42GGATRR531 pKa = 11.84LKK533 pKa = 10.63PEE535 pKa = 4.0VAVSYY540 pKa = 10.81SDD542 pKa = 3.74PSYY545 pKa = 11.02HH546 pKa = 7.24DD547 pKa = 3.68LVEE550 pKa = 3.83RR551 pKa = 11.84TYY553 pKa = 10.69PYY555 pKa = 7.96TTAATFTAVEE565 pKa = 4.17GSITCFMVSGLPQCITLIEE584 pKa = 4.68GDD586 pKa = 3.54SSAYY590 pKa = 8.52ATNLQTYY597 pKa = 7.43NLWSLPIDD605 pKa = 4.1CPSTRR610 pKa = 11.84NLTLTTNTTSPP621 pKa = 3.51
MM1 pKa = 7.32KK2 pKa = 10.29RR3 pKa = 11.84VLDD6 pKa = 3.71WVHH9 pKa = 6.34QKK11 pKa = 7.91MARR14 pKa = 11.84KK15 pKa = 9.49GLDD18 pKa = 3.89PINPTIKK25 pKa = 10.09VGRR28 pKa = 11.84DD29 pKa = 3.2GTEE32 pKa = 3.43VDD34 pKa = 3.44QGLAIYY40 pKa = 9.6IARR43 pKa = 11.84VTVSILAIILLTVLLVTPTVTGLFQVFDD71 pKa = 4.07SSASEE76 pKa = 3.87QSHH79 pKa = 7.37PIMDD83 pKa = 5.11LLNDD87 pKa = 3.66EE88 pKa = 4.7VLPPLRR94 pKa = 11.84NTHH97 pKa = 6.59ALLEE101 pKa = 4.5EE102 pKa = 4.36DD103 pKa = 3.25TAPRR107 pKa = 11.84VRR109 pKa = 11.84SIQTAVEE116 pKa = 3.69LRR118 pKa = 11.84IPGDD122 pKa = 3.61LKK124 pKa = 11.22SATQQILHH132 pKa = 7.31AIKK135 pKa = 9.08DD136 pKa = 3.8TCVGGDD142 pKa = 3.46GGGTGPAPSCPPEE155 pKa = 4.43GVLPQHH161 pKa = 6.72HH162 pKa = 6.63SAFKK166 pKa = 10.47PIDD169 pKa = 4.07PDD171 pKa = 3.78SFTPCATGKK180 pKa = 8.35PTFSGSPAASSSIGSFPWISSGTQDD205 pKa = 3.24TPKK208 pKa = 10.44ISMTMSSSYY217 pKa = 11.01LGFYY221 pKa = 9.57ATDD224 pKa = 3.54TYY226 pKa = 10.71ITTGQNKK233 pKa = 9.61RR234 pKa = 11.84SVSVIGLGRR243 pKa = 11.84LTVTEE248 pKa = 4.85DD249 pKa = 3.27GRR251 pKa = 11.84SSAVVTEE258 pKa = 4.64AEE260 pKa = 4.78PIHH263 pKa = 6.38HH264 pKa = 6.9SSLIGYY270 pKa = 8.77QSTGTGITGTVSFFRR285 pKa = 11.84TTGSEE290 pKa = 4.08DD291 pKa = 3.28TADD294 pKa = 3.82AQDD297 pKa = 3.37VGIRR301 pKa = 11.84SHH303 pKa = 6.24TLLVMRR309 pKa = 11.84RR310 pKa = 11.84NRR312 pKa = 11.84KK313 pKa = 9.08LDD315 pKa = 3.31TYY317 pKa = 10.16VYY319 pKa = 10.32EE320 pKa = 4.54GGEE323 pKa = 3.97IQGGDD328 pKa = 4.39MYY330 pKa = 11.37MMASPLSGQGTMVGDD345 pKa = 3.72EE346 pKa = 4.31MVLLGYY352 pKa = 10.07AVPVTNYY359 pKa = 10.0AGPVWCPNTFCNTQTMEE376 pKa = 3.91ACKK379 pKa = 10.14NASGTIRR386 pKa = 11.84FPGSSTGLAVVMVRR400 pKa = 11.84YY401 pKa = 8.7MIQDD405 pKa = 3.66LDD407 pKa = 3.81KK408 pKa = 11.61DD409 pKa = 3.88GFKK412 pKa = 10.83PKK414 pKa = 9.14ITIHH418 pKa = 5.28MLNRR422 pKa = 11.84EE423 pKa = 4.02QWPIPGPYY431 pKa = 9.71HH432 pKa = 5.82LTSGTKK438 pKa = 9.73GLFISVYY445 pKa = 7.86SWGWYY450 pKa = 6.75GHH452 pKa = 6.1PVSGEE457 pKa = 3.8INRR460 pKa = 11.84ATWDD464 pKa = 3.3MRR466 pKa = 11.84SIRR469 pKa = 11.84LVYY472 pKa = 10.54ALLWPDD478 pKa = 3.48LPEE481 pKa = 4.45TEE483 pKa = 4.5GKK485 pKa = 10.77CYY487 pKa = 10.5TNLANPCPRR496 pKa = 11.84ACTIGSTRR504 pKa = 11.84IGSGIPLTFTGQLSAWLTISADD526 pKa = 3.42GGATRR531 pKa = 11.84LKK533 pKa = 10.63PEE535 pKa = 4.0VAVSYY540 pKa = 10.81SDD542 pKa = 3.74PSYY545 pKa = 11.02HH546 pKa = 7.24DD547 pKa = 3.68LVEE550 pKa = 3.83RR551 pKa = 11.84TYY553 pKa = 10.69PYY555 pKa = 7.96TTAATFTAVEE565 pKa = 4.17GSITCFMVSGLPQCITLIEE584 pKa = 4.68GDD586 pKa = 3.54SSAYY590 pKa = 8.52ATNLQTYY597 pKa = 7.43NLWSLPIDD605 pKa = 4.1CPSTRR610 pKa = 11.84NLTLTTNTTSPP621 pKa = 3.51
Molecular weight: 66.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GMY6|A0A2P1GMY6_9MONO Nucleocapsid protein OS=Wenling triplecross lizardfish paramyxovirus OX=2116451 PE=3 SV=1
MM1 pKa = 7.33SGKK4 pKa = 9.72SAPLWDD10 pKa = 4.45GCWSGGGCRR19 pKa = 11.84DD20 pKa = 3.65NIEE23 pKa = 4.04IPKK26 pKa = 9.85PDD28 pKa = 3.75IEE30 pKa = 4.95KK31 pKa = 10.76GPIPITYY38 pKa = 9.07QVPVPSDD45 pKa = 3.53PGNKK49 pKa = 10.2LITQLTLFGFIEE61 pKa = 4.28YY62 pKa = 10.31KK63 pKa = 10.88YY64 pKa = 9.63MSKK67 pKa = 10.83NSGGEE72 pKa = 3.8KK73 pKa = 9.97KK74 pKa = 10.53GVSRR78 pKa = 11.84IILPCSMPRR87 pKa = 11.84GDD89 pKa = 4.54PDD91 pKa = 3.3IAQMSHH97 pKa = 7.22DD98 pKa = 4.39LSQIEE103 pKa = 3.93ISVIRR108 pKa = 11.84RR109 pKa = 11.84PRR111 pKa = 11.84AIEE114 pKa = 4.29SIGFCADD121 pKa = 3.4SVPDD125 pKa = 3.88SLSEE129 pKa = 4.05HH130 pKa = 6.81LGFLLNGMGFPAYY143 pKa = 9.35KK144 pKa = 10.4VSTGVDD150 pKa = 3.22KK151 pKa = 11.01MPASSKK157 pKa = 10.46YY158 pKa = 10.76KK159 pKa = 9.43FIIWAGVLTALSGPGGYY176 pKa = 9.15VVPIRR181 pKa = 11.84RR182 pKa = 11.84MIGGVAKK189 pKa = 10.25NRR191 pKa = 11.84PSGPEE196 pKa = 3.37VKK198 pKa = 9.63MSLKK202 pKa = 9.08ITLKK206 pKa = 9.94TSFDD210 pKa = 3.45IKK212 pKa = 10.94ALAINAEE219 pKa = 4.17KK220 pKa = 10.63CDD222 pKa = 3.95DD223 pKa = 4.65GGWSTDD229 pKa = 2.89VLLYY233 pKa = 10.42IGTISRR239 pKa = 11.84RR240 pKa = 11.84NGRR243 pKa = 11.84PVDD246 pKa = 3.38TEE248 pKa = 3.97YY249 pKa = 11.56AKK251 pKa = 11.07NFASGLGFKK260 pKa = 10.54LGLGSVGGVSIHH272 pKa = 5.32VHH274 pKa = 4.57ATGLKK279 pKa = 8.82WKK281 pKa = 10.52ASLNKK286 pKa = 8.12TTGGKK291 pKa = 10.23KK292 pKa = 10.19NICFPLAEE300 pKa = 4.26VAPVLNRR307 pKa = 11.84VLACRR312 pKa = 11.84DD313 pKa = 3.28TAIIDD318 pKa = 4.21AEE320 pKa = 4.26ICFQATDD327 pKa = 3.43MRR329 pKa = 11.84DD330 pKa = 3.36VRR332 pKa = 11.84EE333 pKa = 4.29TTSVVHH339 pKa = 6.19TLPKK343 pKa = 10.57DD344 pKa = 3.73VIVVNLSHH352 pKa = 7.14
MM1 pKa = 7.33SGKK4 pKa = 9.72SAPLWDD10 pKa = 4.45GCWSGGGCRR19 pKa = 11.84DD20 pKa = 3.65NIEE23 pKa = 4.04IPKK26 pKa = 9.85PDD28 pKa = 3.75IEE30 pKa = 4.95KK31 pKa = 10.76GPIPITYY38 pKa = 9.07QVPVPSDD45 pKa = 3.53PGNKK49 pKa = 10.2LITQLTLFGFIEE61 pKa = 4.28YY62 pKa = 10.31KK63 pKa = 10.88YY64 pKa = 9.63MSKK67 pKa = 10.83NSGGEE72 pKa = 3.8KK73 pKa = 9.97KK74 pKa = 10.53GVSRR78 pKa = 11.84IILPCSMPRR87 pKa = 11.84GDD89 pKa = 4.54PDD91 pKa = 3.3IAQMSHH97 pKa = 7.22DD98 pKa = 4.39LSQIEE103 pKa = 3.93ISVIRR108 pKa = 11.84RR109 pKa = 11.84PRR111 pKa = 11.84AIEE114 pKa = 4.29SIGFCADD121 pKa = 3.4SVPDD125 pKa = 3.88SLSEE129 pKa = 4.05HH130 pKa = 6.81LGFLLNGMGFPAYY143 pKa = 9.35KK144 pKa = 10.4VSTGVDD150 pKa = 3.22KK151 pKa = 11.01MPASSKK157 pKa = 10.46YY158 pKa = 10.76KK159 pKa = 9.43FIIWAGVLTALSGPGGYY176 pKa = 9.15VVPIRR181 pKa = 11.84RR182 pKa = 11.84MIGGVAKK189 pKa = 10.25NRR191 pKa = 11.84PSGPEE196 pKa = 3.37VKK198 pKa = 9.63MSLKK202 pKa = 9.08ITLKK206 pKa = 9.94TSFDD210 pKa = 3.45IKK212 pKa = 10.94ALAINAEE219 pKa = 4.17KK220 pKa = 10.63CDD222 pKa = 3.95DD223 pKa = 4.65GGWSTDD229 pKa = 2.89VLLYY233 pKa = 10.42IGTISRR239 pKa = 11.84RR240 pKa = 11.84NGRR243 pKa = 11.84PVDD246 pKa = 3.38TEE248 pKa = 3.97YY249 pKa = 11.56AKK251 pKa = 11.07NFASGLGFKK260 pKa = 10.54LGLGSVGGVSIHH272 pKa = 5.32VHH274 pKa = 4.57ATGLKK279 pKa = 8.82WKK281 pKa = 10.52ASLNKK286 pKa = 8.12TTGGKK291 pKa = 10.23KK292 pKa = 10.19NICFPLAEE300 pKa = 4.26VAPVLNRR307 pKa = 11.84VLACRR312 pKa = 11.84DD313 pKa = 3.28TAIIDD318 pKa = 4.21AEE320 pKa = 4.26ICFQATDD327 pKa = 3.43MRR329 pKa = 11.84DD330 pKa = 3.36VRR332 pKa = 11.84EE333 pKa = 4.29TTSVVHH339 pKa = 6.19TLPKK343 pKa = 10.57DD344 pKa = 3.73VIVVNLSHH352 pKa = 7.14
Molecular weight: 37.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5064 |
118 |
2180 |
723.4 |
80.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.385 ± 0.889 | 1.54 ± 0.292 |
6.319 ± 0.465 | 5.569 ± 0.672 |
2.942 ± 0.208 | 6.754 ± 0.776 |
2.31 ± 0.34 | 7.366 ± 0.556 |
4.976 ± 0.514 | 9.36 ± 0.639 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.16 ± 0.229 | 3.752 ± 0.26 |
4.562 ± 0.502 | 3.456 ± 0.437 |
5.766 ± 0.474 | 7.721 ± 0.431 |
6.991 ± 0.747 | 6.062 ± 0.419 |
1.106 ± 0.211 | 2.903 ± 0.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
