
Bat coronavirus Rp/Shaanxi2011
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Sarbecovirus; Severe acute respiratory syndrome-related coronavirus
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
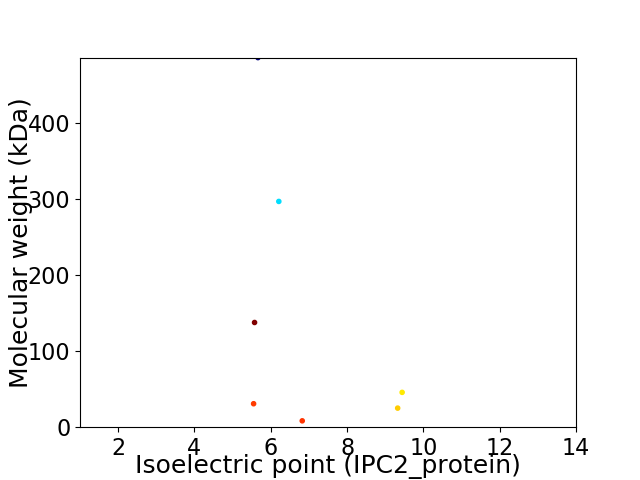
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9QTA1|R9QTA1_SARS Growth factor-like peptide OS=Bat coronavirus Rp/Shaanxi2011 OX=1283332 PE=3 SV=1
MM1 pKa = 7.62ILLLLFLSSAKK12 pKa = 10.29AQEE15 pKa = 4.41GCGVISNKK23 pKa = 7.66PQRR26 pKa = 11.84TFDD29 pKa = 3.5QYY31 pKa = 11.06SSTFRR36 pKa = 11.84GVYY39 pKa = 10.32YY40 pKa = 10.6NDD42 pKa = 4.8DD43 pKa = 3.51IFRR46 pKa = 11.84SDD48 pKa = 3.84VLHH51 pKa = 6.33LTQDD55 pKa = 3.19YY56 pKa = 9.62FLPFNTNVTRR66 pKa = 11.84YY67 pKa = 9.8LSLNAAQNTIVYY79 pKa = 9.09FDD81 pKa = 3.35NHH83 pKa = 7.09VIPFYY88 pKa = 11.04DD89 pKa = 4.89GIYY92 pKa = 9.94FAATEE97 pKa = 4.09RR98 pKa = 11.84SNVIRR103 pKa = 11.84GWIFGSTFDD112 pKa = 4.94NRR114 pKa = 11.84SQSAIIVNNSTHH126 pKa = 6.74ILVKK130 pKa = 9.62VCNFVLCTEE139 pKa = 4.09PMFTVSRR146 pKa = 11.84NQHH149 pKa = 4.75YY150 pKa = 10.34KK151 pKa = 9.14SWVYY155 pKa = 8.83QHH157 pKa = 6.61ARR159 pKa = 11.84NCTYY163 pKa = 11.12DD164 pKa = 3.08VAYY167 pKa = 9.54PSFQLDD173 pKa = 3.13VSLKK177 pKa = 9.87NNVNFQHH184 pKa = 6.09LRR186 pKa = 11.84EE187 pKa = 4.53FIFKK191 pKa = 10.7NVDD194 pKa = 3.09GFLKK198 pKa = 10.13IYY200 pKa = 10.57SSYY203 pKa = 11.11EE204 pKa = 3.83PINVVSGIPSGFSVLKK220 pKa = 10.25PVMSLPLGINITGMRR235 pKa = 11.84VVMTMFSNTQANFLTEE251 pKa = 3.54NAAYY255 pKa = 9.94YY256 pKa = 10.3VGYY259 pKa = 10.29LKK261 pKa = 10.59PRR263 pKa = 11.84TFMLQFNTNGTIVNAVDD280 pKa = 4.46CSQDD284 pKa = 3.36PLSEE288 pKa = 4.17LKK290 pKa = 9.77CTLKK294 pKa = 10.81NFNITKK300 pKa = 10.5GIYY303 pKa = 6.74QTSNFRR309 pKa = 11.84VSPTQEE315 pKa = 3.56VVRR318 pKa = 11.84FPNITNRR325 pKa = 11.84CPFDD329 pKa = 3.51KK330 pKa = 10.89VFNATRR336 pKa = 11.84FPSVYY341 pKa = 9.26AWEE344 pKa = 4.04RR345 pKa = 11.84TKK347 pKa = 10.81ISDD350 pKa = 3.69CVADD354 pKa = 3.7YY355 pKa = 9.98TVLYY359 pKa = 10.29NSTSFSTFKK368 pKa = 10.75CYY370 pKa = 10.03GVSPSKK376 pKa = 11.02LIDD379 pKa = 3.28LCFTSVYY386 pKa = 10.97ADD388 pKa = 3.29TFLIRR393 pKa = 11.84SSEE396 pKa = 4.01VRR398 pKa = 11.84QVAPGEE404 pKa = 4.2TGVIADD410 pKa = 4.42YY411 pKa = 10.82NYY413 pKa = 11.14KK414 pKa = 10.72LPDD417 pKa = 4.27DD418 pKa = 3.81FTGCVIAWNTANQDD432 pKa = 2.97QGQYY436 pKa = 10.33YY437 pKa = 9.01YY438 pKa = 10.87RR439 pKa = 11.84SSRR442 pKa = 11.84KK443 pKa = 9.15EE444 pKa = 3.38KK445 pKa = 10.57LKK447 pKa = 10.65PFEE450 pKa = 5.29RR451 pKa = 11.84DD452 pKa = 3.0LSSDD456 pKa = 3.41EE457 pKa = 4.25NGVYY461 pKa = 9.51TLSTYY466 pKa = 11.13DD467 pKa = 4.25FYY469 pKa = 11.18PSVPLDD475 pKa = 3.6YY476 pKa = 10.54QATRR480 pKa = 11.84VVVLSFEE487 pKa = 4.61LLNAPATVCGPKK499 pKa = 10.49LSTTLVKK506 pKa = 10.42NQCVNFNFNGLKK518 pKa = 9.12GTGVLTASSKK528 pKa = 10.59KK529 pKa = 10.02FQSFQQFGRR538 pKa = 11.84DD539 pKa = 3.18ASDD542 pKa = 3.34FTDD545 pKa = 3.68SVRR548 pKa = 11.84DD549 pKa = 3.77PQTLEE554 pKa = 3.83ILDD557 pKa = 4.16ISPCSFGGVSVITPGTNASTEE578 pKa = 4.27VAVLYY583 pKa = 10.7QDD585 pKa = 4.29VNCTDD590 pKa = 3.07VPTAINADD598 pKa = 3.45QLTPAWRR605 pKa = 11.84VYY607 pKa = 9.55STGINVFQTQAGCLIGAEE625 pKa = 4.44HH626 pKa = 6.46VNASYY631 pKa = 11.03EE632 pKa = 4.01CDD634 pKa = 3.21IPIGAGICASYY645 pKa = 8.41HH646 pKa = 4.65TASVLRR652 pKa = 11.84STGQKK657 pKa = 10.41SIVAYY662 pKa = 9.53TMSLGAEE669 pKa = 3.91NSIAYY674 pKa = 9.44ANNSIAIPTNFSISVTTEE692 pKa = 3.49VMPVSMAKK700 pKa = 9.66TSVDD704 pKa = 3.1CTMYY708 pKa = 10.29ICGDD712 pKa = 3.56SLEE715 pKa = 4.61CSNLLLQYY723 pKa = 11.24GSFCTQLNRR732 pKa = 11.84ALTGIAIEE740 pKa = 4.08QDD742 pKa = 3.23KK743 pKa = 8.9NTQEE747 pKa = 3.75VFAQVKK753 pKa = 9.11QMYY756 pKa = 7.06KK757 pKa = 9.37TPAIKK762 pKa = 10.61DD763 pKa = 3.33FGGFNFSQILPDD775 pKa = 3.66PSKK778 pKa = 8.47PTKK781 pKa = 10.11RR782 pKa = 11.84SFIEE786 pKa = 4.3DD787 pKa = 3.61LLFNKK792 pKa = 7.99VTLADD797 pKa = 3.57AGFMKK802 pKa = 10.37QYY804 pKa = 10.74GEE806 pKa = 4.27CLGDD810 pKa = 3.41ISARR814 pKa = 11.84DD815 pKa = 4.03LICAQKK821 pKa = 10.77FNGLTVLPPLLTDD834 pKa = 3.7EE835 pKa = 5.07MIAAYY840 pKa = 8.13TAALVSGTATAGWTFGAGAALQIPFAMQMAYY871 pKa = 10.08RR872 pKa = 11.84FNGIGVTQNVLYY884 pKa = 10.46EE885 pKa = 4.09NQKK888 pKa = 10.62QIANQFNKK896 pKa = 10.46AISQIQEE903 pKa = 4.19SLTTTSTALGKK914 pKa = 10.37LQDD917 pKa = 3.95VVNQNAQALNTLVKK931 pKa = 10.38QLSSNFGAISSVLNDD946 pKa = 3.17ILSRR950 pKa = 11.84LDD952 pKa = 3.31KK953 pKa = 11.45VEE955 pKa = 5.19AEE957 pKa = 4.28VQIDD961 pKa = 3.56RR962 pKa = 11.84LITGRR967 pKa = 11.84LQSLQTYY974 pKa = 7.4VTQQLIRR981 pKa = 11.84AAEE984 pKa = 3.85IRR986 pKa = 11.84ASANLAATKK995 pKa = 9.96MSEE998 pKa = 4.28CVLGQSKK1005 pKa = 10.12RR1006 pKa = 11.84VDD1008 pKa = 4.0FCGKK1012 pKa = 9.92GYY1014 pKa = 10.57HH1015 pKa = 6.84LMSFPQAAPHH1025 pKa = 5.76GVVFLHH1031 pKa = 5.25VTYY1034 pKa = 10.82VPSQEE1039 pKa = 4.9RR1040 pKa = 11.84NFTTAPAICHH1050 pKa = 5.73EE1051 pKa = 4.45GKK1053 pKa = 10.56AYY1055 pKa = 9.93FPRR1058 pKa = 11.84EE1059 pKa = 3.84GVFVSNGTSWFITQRR1074 pKa = 11.84NFYY1077 pKa = 9.51SPQIITTDD1085 pKa = 3.18NTFVAGSCNVVIGIINNTVYY1105 pKa = 10.97DD1106 pKa = 4.22PLQPEE1111 pKa = 4.19LDD1113 pKa = 3.85SFKK1116 pKa = 11.4EE1117 pKa = 3.91EE1118 pKa = 3.56LDD1120 pKa = 3.63KK1121 pKa = 11.53YY1122 pKa = 10.15FKK1124 pKa = 10.99NHH1126 pKa = 6.01TSPDD1130 pKa = 3.14VDD1132 pKa = 4.4LGDD1135 pKa = 3.31ISGINASVVNIQKK1148 pKa = 10.63EE1149 pKa = 3.92IDD1151 pKa = 3.67RR1152 pKa = 11.84LNEE1155 pKa = 3.5VAKK1158 pKa = 10.61NLNEE1162 pKa = 4.14SLIDD1166 pKa = 3.75LQEE1169 pKa = 4.17LGKK1172 pKa = 10.54YY1173 pKa = 6.98EE1174 pKa = 5.3QYY1176 pKa = 10.78IKK1178 pKa = 10.03WPWYY1182 pKa = 8.39VWLGFIAGLIAIVMVTILLCCMTSCCSCLKK1212 pKa = 10.16GACSCGSCCKK1222 pKa = 10.16FDD1224 pKa = 4.43EE1225 pKa = 5.39DD1226 pKa = 5.38DD1227 pKa = 4.38SEE1229 pKa = 4.36PVLKK1233 pKa = 10.36GVKK1236 pKa = 9.35LHH1238 pKa = 5.3YY1239 pKa = 9.44TT1240 pKa = 3.44
MM1 pKa = 7.62ILLLLFLSSAKK12 pKa = 10.29AQEE15 pKa = 4.41GCGVISNKK23 pKa = 7.66PQRR26 pKa = 11.84TFDD29 pKa = 3.5QYY31 pKa = 11.06SSTFRR36 pKa = 11.84GVYY39 pKa = 10.32YY40 pKa = 10.6NDD42 pKa = 4.8DD43 pKa = 3.51IFRR46 pKa = 11.84SDD48 pKa = 3.84VLHH51 pKa = 6.33LTQDD55 pKa = 3.19YY56 pKa = 9.62FLPFNTNVTRR66 pKa = 11.84YY67 pKa = 9.8LSLNAAQNTIVYY79 pKa = 9.09FDD81 pKa = 3.35NHH83 pKa = 7.09VIPFYY88 pKa = 11.04DD89 pKa = 4.89GIYY92 pKa = 9.94FAATEE97 pKa = 4.09RR98 pKa = 11.84SNVIRR103 pKa = 11.84GWIFGSTFDD112 pKa = 4.94NRR114 pKa = 11.84SQSAIIVNNSTHH126 pKa = 6.74ILVKK130 pKa = 9.62VCNFVLCTEE139 pKa = 4.09PMFTVSRR146 pKa = 11.84NQHH149 pKa = 4.75YY150 pKa = 10.34KK151 pKa = 9.14SWVYY155 pKa = 8.83QHH157 pKa = 6.61ARR159 pKa = 11.84NCTYY163 pKa = 11.12DD164 pKa = 3.08VAYY167 pKa = 9.54PSFQLDD173 pKa = 3.13VSLKK177 pKa = 9.87NNVNFQHH184 pKa = 6.09LRR186 pKa = 11.84EE187 pKa = 4.53FIFKK191 pKa = 10.7NVDD194 pKa = 3.09GFLKK198 pKa = 10.13IYY200 pKa = 10.57SSYY203 pKa = 11.11EE204 pKa = 3.83PINVVSGIPSGFSVLKK220 pKa = 10.25PVMSLPLGINITGMRR235 pKa = 11.84VVMTMFSNTQANFLTEE251 pKa = 3.54NAAYY255 pKa = 9.94YY256 pKa = 10.3VGYY259 pKa = 10.29LKK261 pKa = 10.59PRR263 pKa = 11.84TFMLQFNTNGTIVNAVDD280 pKa = 4.46CSQDD284 pKa = 3.36PLSEE288 pKa = 4.17LKK290 pKa = 9.77CTLKK294 pKa = 10.81NFNITKK300 pKa = 10.5GIYY303 pKa = 6.74QTSNFRR309 pKa = 11.84VSPTQEE315 pKa = 3.56VVRR318 pKa = 11.84FPNITNRR325 pKa = 11.84CPFDD329 pKa = 3.51KK330 pKa = 10.89VFNATRR336 pKa = 11.84FPSVYY341 pKa = 9.26AWEE344 pKa = 4.04RR345 pKa = 11.84TKK347 pKa = 10.81ISDD350 pKa = 3.69CVADD354 pKa = 3.7YY355 pKa = 9.98TVLYY359 pKa = 10.29NSTSFSTFKK368 pKa = 10.75CYY370 pKa = 10.03GVSPSKK376 pKa = 11.02LIDD379 pKa = 3.28LCFTSVYY386 pKa = 10.97ADD388 pKa = 3.29TFLIRR393 pKa = 11.84SSEE396 pKa = 4.01VRR398 pKa = 11.84QVAPGEE404 pKa = 4.2TGVIADD410 pKa = 4.42YY411 pKa = 10.82NYY413 pKa = 11.14KK414 pKa = 10.72LPDD417 pKa = 4.27DD418 pKa = 3.81FTGCVIAWNTANQDD432 pKa = 2.97QGQYY436 pKa = 10.33YY437 pKa = 9.01YY438 pKa = 10.87RR439 pKa = 11.84SSRR442 pKa = 11.84KK443 pKa = 9.15EE444 pKa = 3.38KK445 pKa = 10.57LKK447 pKa = 10.65PFEE450 pKa = 5.29RR451 pKa = 11.84DD452 pKa = 3.0LSSDD456 pKa = 3.41EE457 pKa = 4.25NGVYY461 pKa = 9.51TLSTYY466 pKa = 11.13DD467 pKa = 4.25FYY469 pKa = 11.18PSVPLDD475 pKa = 3.6YY476 pKa = 10.54QATRR480 pKa = 11.84VVVLSFEE487 pKa = 4.61LLNAPATVCGPKK499 pKa = 10.49LSTTLVKK506 pKa = 10.42NQCVNFNFNGLKK518 pKa = 9.12GTGVLTASSKK528 pKa = 10.59KK529 pKa = 10.02FQSFQQFGRR538 pKa = 11.84DD539 pKa = 3.18ASDD542 pKa = 3.34FTDD545 pKa = 3.68SVRR548 pKa = 11.84DD549 pKa = 3.77PQTLEE554 pKa = 3.83ILDD557 pKa = 4.16ISPCSFGGVSVITPGTNASTEE578 pKa = 4.27VAVLYY583 pKa = 10.7QDD585 pKa = 4.29VNCTDD590 pKa = 3.07VPTAINADD598 pKa = 3.45QLTPAWRR605 pKa = 11.84VYY607 pKa = 9.55STGINVFQTQAGCLIGAEE625 pKa = 4.44HH626 pKa = 6.46VNASYY631 pKa = 11.03EE632 pKa = 4.01CDD634 pKa = 3.21IPIGAGICASYY645 pKa = 8.41HH646 pKa = 4.65TASVLRR652 pKa = 11.84STGQKK657 pKa = 10.41SIVAYY662 pKa = 9.53TMSLGAEE669 pKa = 3.91NSIAYY674 pKa = 9.44ANNSIAIPTNFSISVTTEE692 pKa = 3.49VMPVSMAKK700 pKa = 9.66TSVDD704 pKa = 3.1CTMYY708 pKa = 10.29ICGDD712 pKa = 3.56SLEE715 pKa = 4.61CSNLLLQYY723 pKa = 11.24GSFCTQLNRR732 pKa = 11.84ALTGIAIEE740 pKa = 4.08QDD742 pKa = 3.23KK743 pKa = 8.9NTQEE747 pKa = 3.75VFAQVKK753 pKa = 9.11QMYY756 pKa = 7.06KK757 pKa = 9.37TPAIKK762 pKa = 10.61DD763 pKa = 3.33FGGFNFSQILPDD775 pKa = 3.66PSKK778 pKa = 8.47PTKK781 pKa = 10.11RR782 pKa = 11.84SFIEE786 pKa = 4.3DD787 pKa = 3.61LLFNKK792 pKa = 7.99VTLADD797 pKa = 3.57AGFMKK802 pKa = 10.37QYY804 pKa = 10.74GEE806 pKa = 4.27CLGDD810 pKa = 3.41ISARR814 pKa = 11.84DD815 pKa = 4.03LICAQKK821 pKa = 10.77FNGLTVLPPLLTDD834 pKa = 3.7EE835 pKa = 5.07MIAAYY840 pKa = 8.13TAALVSGTATAGWTFGAGAALQIPFAMQMAYY871 pKa = 10.08RR872 pKa = 11.84FNGIGVTQNVLYY884 pKa = 10.46EE885 pKa = 4.09NQKK888 pKa = 10.62QIANQFNKK896 pKa = 10.46AISQIQEE903 pKa = 4.19SLTTTSTALGKK914 pKa = 10.37LQDD917 pKa = 3.95VVNQNAQALNTLVKK931 pKa = 10.38QLSSNFGAISSVLNDD946 pKa = 3.17ILSRR950 pKa = 11.84LDD952 pKa = 3.31KK953 pKa = 11.45VEE955 pKa = 5.19AEE957 pKa = 4.28VQIDD961 pKa = 3.56RR962 pKa = 11.84LITGRR967 pKa = 11.84LQSLQTYY974 pKa = 7.4VTQQLIRR981 pKa = 11.84AAEE984 pKa = 3.85IRR986 pKa = 11.84ASANLAATKK995 pKa = 9.96MSEE998 pKa = 4.28CVLGQSKK1005 pKa = 10.12RR1006 pKa = 11.84VDD1008 pKa = 4.0FCGKK1012 pKa = 9.92GYY1014 pKa = 10.57HH1015 pKa = 6.84LMSFPQAAPHH1025 pKa = 5.76GVVFLHH1031 pKa = 5.25VTYY1034 pKa = 10.82VPSQEE1039 pKa = 4.9RR1040 pKa = 11.84NFTTAPAICHH1050 pKa = 5.73EE1051 pKa = 4.45GKK1053 pKa = 10.56AYY1055 pKa = 9.93FPRR1058 pKa = 11.84EE1059 pKa = 3.84GVFVSNGTSWFITQRR1074 pKa = 11.84NFYY1077 pKa = 9.51SPQIITTDD1085 pKa = 3.18NTFVAGSCNVVIGIINNTVYY1105 pKa = 10.97DD1106 pKa = 4.22PLQPEE1111 pKa = 4.19LDD1113 pKa = 3.85SFKK1116 pKa = 11.4EE1117 pKa = 3.91EE1118 pKa = 3.56LDD1120 pKa = 3.63KK1121 pKa = 11.53YY1122 pKa = 10.15FKK1124 pKa = 10.99NHH1126 pKa = 6.01TSPDD1130 pKa = 3.14VDD1132 pKa = 4.4LGDD1135 pKa = 3.31ISGINASVVNIQKK1148 pKa = 10.63EE1149 pKa = 3.92IDD1151 pKa = 3.67RR1152 pKa = 11.84LNEE1155 pKa = 3.5VAKK1158 pKa = 10.61NLNEE1162 pKa = 4.14SLIDD1166 pKa = 3.75LQEE1169 pKa = 4.17LGKK1172 pKa = 10.54YY1173 pKa = 6.98EE1174 pKa = 5.3QYY1176 pKa = 10.78IKK1178 pKa = 10.03WPWYY1182 pKa = 8.39VWLGFIAGLIAIVMVTILLCCMTSCCSCLKK1212 pKa = 10.16GACSCGSCCKK1222 pKa = 10.16FDD1224 pKa = 4.43EE1225 pKa = 5.39DD1226 pKa = 5.38DD1227 pKa = 4.38SEE1229 pKa = 4.36PVLKK1233 pKa = 10.36GVKK1236 pKa = 9.35LHH1238 pKa = 5.3YY1239 pKa = 9.44TT1240 pKa = 3.44
Molecular weight: 137.63 kDa
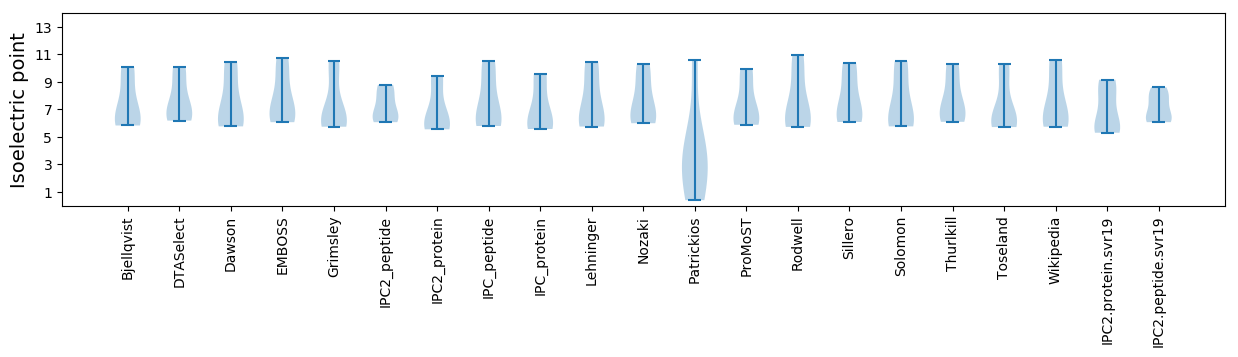
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9QTH1|R9QTH1_SARS ORF3 protein OS=Bat coronavirus Rp/Shaanxi2011 OX=1283332 PE=4 SV=1
MM1 pKa = 7.77SDD3 pKa = 4.56NGPQNQRR10 pKa = 11.84SAPRR14 pKa = 11.84ITFGGPSDD22 pKa = 3.8STDD25 pKa = 3.04NNQDD29 pKa = 3.07GGRR32 pKa = 11.84SGARR36 pKa = 11.84PKK38 pKa = 10.25QRR40 pKa = 11.84RR41 pKa = 11.84PQGLPNNTASWFTALTQHH59 pKa = 6.06GKK61 pKa = 10.36EE62 pKa = 3.96EE63 pKa = 4.07LRR65 pKa = 11.84FPRR68 pKa = 11.84GQGVPINTNSGKK80 pKa = 10.01DD81 pKa = 3.58DD82 pKa = 3.56QIGYY86 pKa = 8.17YY87 pKa = 9.92RR88 pKa = 11.84RR89 pKa = 11.84ATRR92 pKa = 11.84RR93 pKa = 11.84VRR95 pKa = 11.84GGDD98 pKa = 3.09GKK100 pKa = 10.39MKK102 pKa = 9.92EE103 pKa = 4.5LSPRR107 pKa = 11.84WYY109 pKa = 10.01FYY111 pKa = 11.5YY112 pKa = 10.73LGTGPEE118 pKa = 3.63ASLPYY123 pKa = 9.9GANKK127 pKa = 10.09EE128 pKa = 4.16GIIWVATEE136 pKa = 4.18GALNTPKK143 pKa = 10.91DD144 pKa = 4.0HH145 pKa = 7.46IGTRR149 pKa = 11.84NPNNNAAIVLQLPQGTTLPKK169 pKa = 10.34GFYY172 pKa = 10.67AEE174 pKa = 4.5GSRR177 pKa = 11.84GGSQASSRR185 pKa = 11.84SSSRR189 pKa = 11.84SRR191 pKa = 11.84GNSRR195 pKa = 11.84NSTPGSSRR203 pKa = 11.84GNSPARR209 pKa = 11.84MASGGGEE216 pKa = 3.84TALALLLLDD225 pKa = 4.89RR226 pKa = 11.84LNQLEE231 pKa = 4.58SKK233 pKa = 10.86VSGKK237 pKa = 10.04GQQQQGQTVTKK248 pKa = 9.98KK249 pKa = 10.44SAAEE253 pKa = 3.73ASKK256 pKa = 10.76KK257 pKa = 9.61PRR259 pKa = 11.84QKK261 pKa = 9.55RR262 pKa = 11.84TATKK266 pKa = 10.22SYY268 pKa = 11.04NVTQAFGRR276 pKa = 11.84RR277 pKa = 11.84GPEE280 pKa = 3.4QTQGNFGDD288 pKa = 3.74QDD290 pKa = 5.15LIRR293 pKa = 11.84QGTDD297 pKa = 2.96YY298 pKa = 11.27KK299 pKa = 10.5HH300 pKa = 6.32WPQIAQFAPSASAFFGMSRR319 pKa = 11.84IGMEE323 pKa = 4.16VTPSGTWLTYY333 pKa = 10.42HH334 pKa = 7.06GAIKK338 pKa = 10.61LDD340 pKa = 4.18DD341 pKa = 4.89KK342 pKa = 11.64DD343 pKa = 3.91PQFKK347 pKa = 11.14DD348 pKa = 3.7NVILLNKK355 pKa = 10.01HH356 pKa = 4.67IDD358 pKa = 3.35AYY360 pKa = 9.77KK361 pKa = 9.88TFPPTEE367 pKa = 3.95PKK369 pKa = 9.88KK370 pKa = 10.6DD371 pKa = 3.48KK372 pKa = 10.73KK373 pKa = 10.99KK374 pKa = 9.88KK375 pKa = 8.29TDD377 pKa = 3.29EE378 pKa = 4.34AQPLPQRR385 pKa = 11.84KK386 pKa = 8.41KK387 pKa = 10.06QPTVTLLPAADD398 pKa = 3.84MDD400 pKa = 4.26DD401 pKa = 5.3FSRR404 pKa = 11.84QLQNSMSGASADD416 pKa = 3.63STQAA420 pKa = 3.04
MM1 pKa = 7.77SDD3 pKa = 4.56NGPQNQRR10 pKa = 11.84SAPRR14 pKa = 11.84ITFGGPSDD22 pKa = 3.8STDD25 pKa = 3.04NNQDD29 pKa = 3.07GGRR32 pKa = 11.84SGARR36 pKa = 11.84PKK38 pKa = 10.25QRR40 pKa = 11.84RR41 pKa = 11.84PQGLPNNTASWFTALTQHH59 pKa = 6.06GKK61 pKa = 10.36EE62 pKa = 3.96EE63 pKa = 4.07LRR65 pKa = 11.84FPRR68 pKa = 11.84GQGVPINTNSGKK80 pKa = 10.01DD81 pKa = 3.58DD82 pKa = 3.56QIGYY86 pKa = 8.17YY87 pKa = 9.92RR88 pKa = 11.84RR89 pKa = 11.84ATRR92 pKa = 11.84RR93 pKa = 11.84VRR95 pKa = 11.84GGDD98 pKa = 3.09GKK100 pKa = 10.39MKK102 pKa = 9.92EE103 pKa = 4.5LSPRR107 pKa = 11.84WYY109 pKa = 10.01FYY111 pKa = 11.5YY112 pKa = 10.73LGTGPEE118 pKa = 3.63ASLPYY123 pKa = 9.9GANKK127 pKa = 10.09EE128 pKa = 4.16GIIWVATEE136 pKa = 4.18GALNTPKK143 pKa = 10.91DD144 pKa = 4.0HH145 pKa = 7.46IGTRR149 pKa = 11.84NPNNNAAIVLQLPQGTTLPKK169 pKa = 10.34GFYY172 pKa = 10.67AEE174 pKa = 4.5GSRR177 pKa = 11.84GGSQASSRR185 pKa = 11.84SSSRR189 pKa = 11.84SRR191 pKa = 11.84GNSRR195 pKa = 11.84NSTPGSSRR203 pKa = 11.84GNSPARR209 pKa = 11.84MASGGGEE216 pKa = 3.84TALALLLLDD225 pKa = 4.89RR226 pKa = 11.84LNQLEE231 pKa = 4.58SKK233 pKa = 10.86VSGKK237 pKa = 10.04GQQQQGQTVTKK248 pKa = 9.98KK249 pKa = 10.44SAAEE253 pKa = 3.73ASKK256 pKa = 10.76KK257 pKa = 9.61PRR259 pKa = 11.84QKK261 pKa = 9.55RR262 pKa = 11.84TATKK266 pKa = 10.22SYY268 pKa = 11.04NVTQAFGRR276 pKa = 11.84RR277 pKa = 11.84GPEE280 pKa = 3.4QTQGNFGDD288 pKa = 3.74QDD290 pKa = 5.15LIRR293 pKa = 11.84QGTDD297 pKa = 2.96YY298 pKa = 11.27KK299 pKa = 10.5HH300 pKa = 6.32WPQIAQFAPSASAFFGMSRR319 pKa = 11.84IGMEE323 pKa = 4.16VTPSGTWLTYY333 pKa = 10.42HH334 pKa = 7.06GAIKK338 pKa = 10.61LDD340 pKa = 4.18DD341 pKa = 4.89KK342 pKa = 11.64DD343 pKa = 3.91PQFKK347 pKa = 11.14DD348 pKa = 3.7NVILLNKK355 pKa = 10.01HH356 pKa = 4.67IDD358 pKa = 3.35AYY360 pKa = 9.77KK361 pKa = 9.88TFPPTEE367 pKa = 3.95PKK369 pKa = 9.88KK370 pKa = 10.6DD371 pKa = 3.48KK372 pKa = 10.73KK373 pKa = 10.99KK374 pKa = 9.88KK375 pKa = 8.29TDD377 pKa = 3.29EE378 pKa = 4.34AQPLPQRR385 pKa = 11.84KK386 pKa = 8.41KK387 pKa = 10.06QPTVTLLPAADD398 pKa = 3.84MDD400 pKa = 4.26DD401 pKa = 5.3FSRR404 pKa = 11.84QLQNSMSGASADD416 pKa = 3.63STQAA420 pKa = 3.04
Molecular weight: 45.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9233 |
76 |
4374 |
1319.0 |
147.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.181 ± 0.157 | 3.033 ± 0.403 |
5.22 ± 0.496 | 4.679 ± 0.638 |
4.787 ± 0.337 | 6.076 ± 0.487 |
2.015 ± 0.314 | 5.058 ± 0.614 |
5.675 ± 0.51 | 9.412 ± 0.65 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.212 | 5.318 ± 0.382 |
4.04 ± 0.316 | 3.78 ± 0.515 |
3.737 ± 0.504 | 6.867 ± 0.447 |
7.062 ± 0.22 | 8.036 ± 0.635 |
1.137 ± 0.222 | 4.56 ± 0.294 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
