
Gloeocapsa sp. PCC 73106
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Chroococcaceae; Gloeocapsa; unclassified Gloeocapsa
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
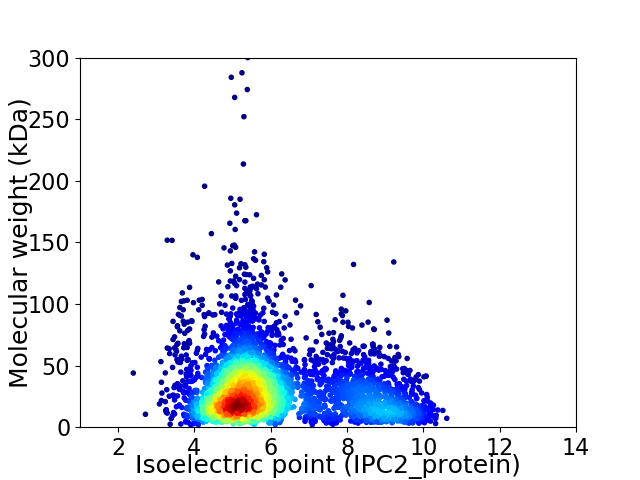
Virtual 2D-PAGE plot for 4062 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8LR29|L8LR29_9CHRO Uncharacterized protein OS=Gloeocapsa sp. PCC 73106 OX=102232 GN=GLO73106DRAFT_00014570 PE=4 SV=1
GG1 pKa = 7.54DD2 pKa = 3.73APAPVVGLINQYY14 pKa = 9.78GNRR17 pKa = 11.84GQNIPWNIAIANDD30 pKa = 3.76DD31 pKa = 3.92SGDD34 pKa = 3.6VYY36 pKa = 11.25ATGTTAGNVGGPSAGSLDD54 pKa = 3.97GYY56 pKa = 10.74LLQFNQIGVRR66 pKa = 11.84GWSKK70 pKa = 10.79QLGTTEE76 pKa = 3.64IDD78 pKa = 3.13NIYY81 pKa = 10.55GVNTNTDD88 pKa = 2.64GDD90 pKa = 4.41VYY92 pKa = 10.87FVGRR96 pKa = 11.84TKK98 pKa = 11.06GSLVAPNAGIGNDD111 pKa = 3.24IVFGKK116 pKa = 10.51YY117 pKa = 10.22DD118 pKa = 3.58SDD120 pKa = 5.5GNEE123 pKa = 3.37QWIQQFGSFGVDD135 pKa = 2.79NSFVNPEE142 pKa = 4.05LDD144 pKa = 2.95ASGNVYY150 pKa = 10.55VGGYY154 pKa = 8.57TDD156 pKa = 4.62GSLFGPNAGAAFPPSLDD173 pKa = 2.98PWVAKK178 pKa = 10.29FDD180 pKa = 4.37PDD182 pKa = 5.29GNQLWARR189 pKa = 11.84QFGSPTGDD197 pKa = 3.05EE198 pKa = 4.63LFGLDD203 pKa = 4.12VDD205 pKa = 4.23SQGNVLATGWTFGGLAGPNNVNVDD229 pKa = 3.34GAATYY234 pKa = 9.65DD235 pKa = 2.82IWLQKK240 pKa = 9.91MDD242 pKa = 4.11TNGNTLWTEE251 pKa = 3.91QFGTNVDD258 pKa = 3.06DD259 pKa = 4.13WAWDD263 pKa = 3.33IEE265 pKa = 4.44TNAAGEE271 pKa = 4.27IYY273 pKa = 9.74LTGWTLGNLGGTNAGSYY290 pKa = 10.4DD291 pKa = 4.07YY292 pKa = 11.16YY293 pKa = 9.47IAKK296 pKa = 10.01YY297 pKa = 10.26SSSGDD302 pKa = 3.69QLWVRR307 pKa = 11.84QYY309 pKa = 10.46GTTGDD314 pKa = 3.92DD315 pKa = 3.2AATRR319 pKa = 11.84MALDD323 pKa = 4.15DD324 pKa = 4.42NGNIYY329 pKa = 8.91LTGYY333 pKa = 10.69SNGPASGQVFGTSGGYY349 pKa = 9.3DD350 pKa = 2.7AWVAKK355 pKa = 10.1FDD357 pKa = 4.64SEE359 pKa = 5.21GNSIWRR365 pKa = 11.84KK366 pKa = 7.58TIGTVEE372 pKa = 4.28LDD374 pKa = 3.35QAFGIAVQGNDD385 pKa = 3.92LLVSGLTEE393 pKa = 4.31GSLGAVNQGSYY404 pKa = 10.77DD405 pKa = 3.15GWIARR410 pKa = 11.84LTTTTGNLLSFNGLGG425 pKa = 3.38
GG1 pKa = 7.54DD2 pKa = 3.73APAPVVGLINQYY14 pKa = 9.78GNRR17 pKa = 11.84GQNIPWNIAIANDD30 pKa = 3.76DD31 pKa = 3.92SGDD34 pKa = 3.6VYY36 pKa = 11.25ATGTTAGNVGGPSAGSLDD54 pKa = 3.97GYY56 pKa = 10.74LLQFNQIGVRR66 pKa = 11.84GWSKK70 pKa = 10.79QLGTTEE76 pKa = 3.64IDD78 pKa = 3.13NIYY81 pKa = 10.55GVNTNTDD88 pKa = 2.64GDD90 pKa = 4.41VYY92 pKa = 10.87FVGRR96 pKa = 11.84TKK98 pKa = 11.06GSLVAPNAGIGNDD111 pKa = 3.24IVFGKK116 pKa = 10.51YY117 pKa = 10.22DD118 pKa = 3.58SDD120 pKa = 5.5GNEE123 pKa = 3.37QWIQQFGSFGVDD135 pKa = 2.79NSFVNPEE142 pKa = 4.05LDD144 pKa = 2.95ASGNVYY150 pKa = 10.55VGGYY154 pKa = 8.57TDD156 pKa = 4.62GSLFGPNAGAAFPPSLDD173 pKa = 2.98PWVAKK178 pKa = 10.29FDD180 pKa = 4.37PDD182 pKa = 5.29GNQLWARR189 pKa = 11.84QFGSPTGDD197 pKa = 3.05EE198 pKa = 4.63LFGLDD203 pKa = 4.12VDD205 pKa = 4.23SQGNVLATGWTFGGLAGPNNVNVDD229 pKa = 3.34GAATYY234 pKa = 9.65DD235 pKa = 2.82IWLQKK240 pKa = 9.91MDD242 pKa = 4.11TNGNTLWTEE251 pKa = 3.91QFGTNVDD258 pKa = 3.06DD259 pKa = 4.13WAWDD263 pKa = 3.33IEE265 pKa = 4.44TNAAGEE271 pKa = 4.27IYY273 pKa = 9.74LTGWTLGNLGGTNAGSYY290 pKa = 10.4DD291 pKa = 4.07YY292 pKa = 11.16YY293 pKa = 9.47IAKK296 pKa = 10.01YY297 pKa = 10.26SSSGDD302 pKa = 3.69QLWVRR307 pKa = 11.84QYY309 pKa = 10.46GTTGDD314 pKa = 3.92DD315 pKa = 3.2AATRR319 pKa = 11.84MALDD323 pKa = 4.15DD324 pKa = 4.42NGNIYY329 pKa = 8.91LTGYY333 pKa = 10.69SNGPASGQVFGTSGGYY349 pKa = 9.3DD350 pKa = 2.7AWVAKK355 pKa = 10.1FDD357 pKa = 4.64SEE359 pKa = 5.21GNSIWRR365 pKa = 11.84KK366 pKa = 7.58TIGTVEE372 pKa = 4.28LDD374 pKa = 3.35QAFGIAVQGNDD385 pKa = 3.92LLVSGLTEE393 pKa = 4.31GSLGAVNQGSYY404 pKa = 10.77DD405 pKa = 3.15GWIARR410 pKa = 11.84LTTTTGNLLSFNGLGG425 pKa = 3.38
Molecular weight: 44.75 kDa
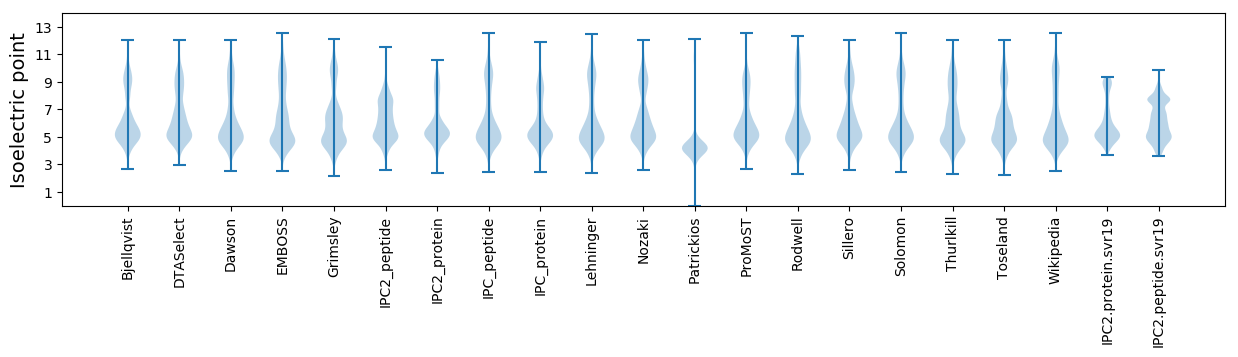
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8LMR8|L8LMR8_9CHRO Transketolase OS=Gloeocapsa sp. PCC 73106 OX=102232 GN=GLO73106DRAFT_00026810 PE=4 SV=1
MM1 pKa = 7.36ATNDD5 pKa = 4.48TISDD9 pKa = 3.51MLTRR13 pKa = 11.84IRR15 pKa = 11.84NACLVRR21 pKa = 11.84QSSTHH26 pKa = 5.07VPTTRR31 pKa = 11.84MTRR34 pKa = 11.84NIARR38 pKa = 11.84VLKK41 pKa = 10.35EE42 pKa = 3.62EE43 pKa = 4.29GFIEE47 pKa = 4.29DD48 pKa = 3.71FQEE51 pKa = 4.2IGEE54 pKa = 4.51GIKK57 pKa = 10.32RR58 pKa = 11.84QLVISLRR65 pKa = 11.84YY66 pKa = 8.71KK67 pKa = 10.69GKK69 pKa = 10.14AGRR72 pKa = 11.84PIIRR76 pKa = 11.84TLKK79 pKa = 10.06RR80 pKa = 11.84VSKK83 pKa = 10.24PGLRR87 pKa = 11.84VYY89 pKa = 9.37TNRR92 pKa = 11.84KK93 pKa = 7.66EE94 pKa = 4.01MPRR97 pKa = 11.84VLGGIGIAIISTSQGIMTDD116 pKa = 3.34RR117 pKa = 11.84EE118 pKa = 4.08ARR120 pKa = 11.84RR121 pKa = 11.84QGIGGEE127 pKa = 3.97ILCYY131 pKa = 10.3VWW133 pKa = 4.93
MM1 pKa = 7.36ATNDD5 pKa = 4.48TISDD9 pKa = 3.51MLTRR13 pKa = 11.84IRR15 pKa = 11.84NACLVRR21 pKa = 11.84QSSTHH26 pKa = 5.07VPTTRR31 pKa = 11.84MTRR34 pKa = 11.84NIARR38 pKa = 11.84VLKK41 pKa = 10.35EE42 pKa = 3.62EE43 pKa = 4.29GFIEE47 pKa = 4.29DD48 pKa = 3.71FQEE51 pKa = 4.2IGEE54 pKa = 4.51GIKK57 pKa = 10.32RR58 pKa = 11.84QLVISLRR65 pKa = 11.84YY66 pKa = 8.71KK67 pKa = 10.69GKK69 pKa = 10.14AGRR72 pKa = 11.84PIIRR76 pKa = 11.84TLKK79 pKa = 10.06RR80 pKa = 11.84VSKK83 pKa = 10.24PGLRR87 pKa = 11.84VYY89 pKa = 9.37TNRR92 pKa = 11.84KK93 pKa = 7.66EE94 pKa = 4.01MPRR97 pKa = 11.84VLGGIGIAIISTSQGIMTDD116 pKa = 3.34RR117 pKa = 11.84EE118 pKa = 4.08ARR120 pKa = 11.84RR121 pKa = 11.84QGIGGEE127 pKa = 3.97ILCYY131 pKa = 10.3VWW133 pKa = 4.93
Molecular weight: 15.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1199467 |
20 |
2695 |
295.3 |
32.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.43 ± 0.042 | 0.927 ± 0.013 |
4.878 ± 0.035 | 6.542 ± 0.038 |
3.98 ± 0.028 | 6.877 ± 0.059 |
1.723 ± 0.019 | 7.38 ± 0.032 |
4.659 ± 0.042 | 11.438 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.736 ± 0.015 | 4.523 ± 0.036 |
4.604 ± 0.03 | 5.199 ± 0.038 |
4.858 ± 0.029 | 6.52 ± 0.032 |
5.773 ± 0.029 | 6.331 ± 0.032 |
1.415 ± 0.021 | 3.208 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
