
Actinobacteria bacterium IMCC26207
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; unclassified Actinomycetia
Average proteome isoelectric point is 5.59
Get precalculated fractions of proteins
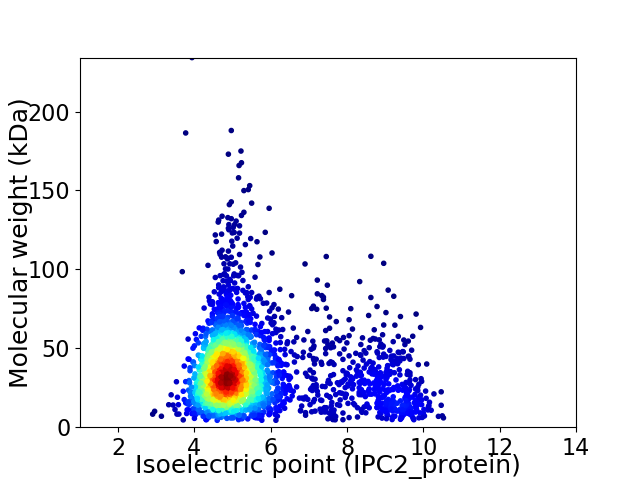
Virtual 2D-PAGE plot for 2975 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
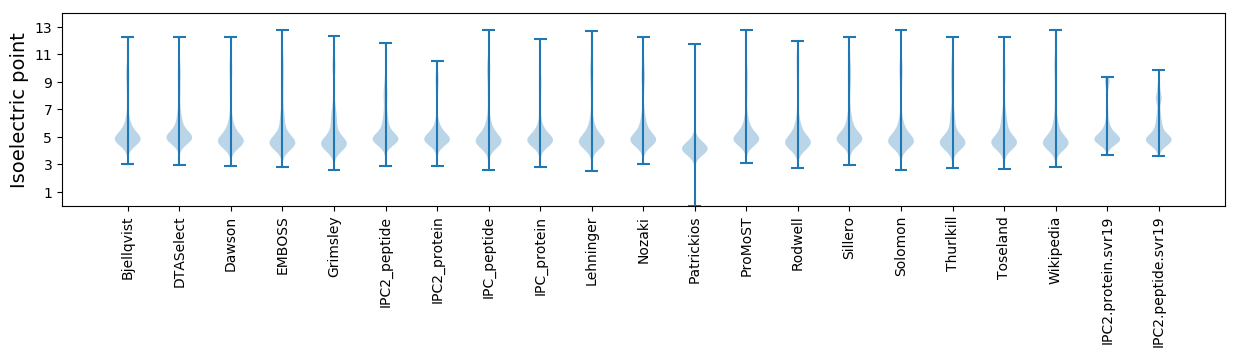
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0J0UXG5|A0A0J0UXG5_9ACTN Methylmalonyl-CoA epimerase OS=Actinobacteria bacterium IMCC26207 OX=1641811 GN=IMCC26207_10743 PE=3 SV=1
MM1 pKa = 7.26KK2 pKa = 10.13NAYY5 pKa = 9.15RR6 pKa = 11.84VLPVVSAALLLFAVSCAPAPGGGGTANLAPIAVATASPLSGNAPLSAAFDD56 pKa = 4.03GSGSVDD62 pKa = 3.08SDD64 pKa = 3.67GSIVSYY70 pKa = 10.4SWSFGNGTNGSGVTSTGNYY89 pKa = 5.27PTAGAFTAVLTVTDD103 pKa = 4.2NKK105 pKa = 10.94GATDD109 pKa = 3.85TDD111 pKa = 4.02SLTITVNGDD120 pKa = 2.91GDD122 pKa = 4.14GDD124 pKa = 4.4GVFVPSDD131 pKa = 3.71CNDD134 pKa = 3.23ADD136 pKa = 3.92GAIFPGAADD145 pKa = 3.64PAGDD149 pKa = 4.78NIDD152 pKa = 4.34SNCDD156 pKa = 3.59GIDD159 pKa = 3.63GTEE162 pKa = 3.94SAAVFVNSSTGANTSTCGTILEE184 pKa = 4.52PCASITQGQSRR195 pKa = 11.84AVAEE199 pKa = 4.55SKK201 pKa = 10.93SSVFVAGGSYY211 pKa = 11.02SKK213 pKa = 10.18FTVQAGLEE221 pKa = 4.06VRR223 pKa = 11.84GGYY226 pKa = 8.54GQNFQRR232 pKa = 11.84GVLATSPSIATVTAAFDD249 pKa = 3.5ATVNGPVGVIADD261 pKa = 5.29GISTATRR268 pKa = 11.84VADD271 pKa = 3.92LSIVGTTASAGSSSYY286 pKa = 11.38GVFVRR291 pKa = 11.84NSTNALVLDD300 pKa = 4.18SLNIAGGVAGAGSAGAAGSSGWSGTAGTGSNGGDD334 pKa = 3.2GFEE337 pKa = 5.28PGGACNTSSAGGGGAGGGGANSGGSGGKK365 pKa = 10.24GGVVDD370 pKa = 6.53GSCGWTGLCSNCDD383 pKa = 3.42AQTGNSGGSGGGVGGGGGGAAGSAATDD410 pKa = 3.79GLVGICTLGGTGPTLRR426 pKa = 11.84GGSGSSGANGTAGGAGSGGSAGANGATGLLGANGAGGGAGGGGGANDD473 pKa = 4.97CNIDD477 pKa = 3.63DD478 pKa = 4.77AGAGGGGGGGGGTRR492 pKa = 11.84ATAGGAGGGAGAQSIGLRR510 pKa = 11.84LQNASPTLIAIQIQLGTGGAGGTGGSGAAGQPGGSGGSGGVAFDD554 pKa = 3.9RR555 pKa = 11.84GGGGGNGGTGGTGGASGAGGGGGGGAAIGLSRR587 pKa = 11.84DD588 pKa = 3.52GSSTPSGSPSYY599 pKa = 10.73SGGVGGSGGSAGSGGNPGATGLVTNTTVAA628 pKa = 3.31
MM1 pKa = 7.26KK2 pKa = 10.13NAYY5 pKa = 9.15RR6 pKa = 11.84VLPVVSAALLLFAVSCAPAPGGGGTANLAPIAVATASPLSGNAPLSAAFDD56 pKa = 4.03GSGSVDD62 pKa = 3.08SDD64 pKa = 3.67GSIVSYY70 pKa = 10.4SWSFGNGTNGSGVTSTGNYY89 pKa = 5.27PTAGAFTAVLTVTDD103 pKa = 4.2NKK105 pKa = 10.94GATDD109 pKa = 3.85TDD111 pKa = 4.02SLTITVNGDD120 pKa = 2.91GDD122 pKa = 4.14GDD124 pKa = 4.4GVFVPSDD131 pKa = 3.71CNDD134 pKa = 3.23ADD136 pKa = 3.92GAIFPGAADD145 pKa = 3.64PAGDD149 pKa = 4.78NIDD152 pKa = 4.34SNCDD156 pKa = 3.59GIDD159 pKa = 3.63GTEE162 pKa = 3.94SAAVFVNSSTGANTSTCGTILEE184 pKa = 4.52PCASITQGQSRR195 pKa = 11.84AVAEE199 pKa = 4.55SKK201 pKa = 10.93SSVFVAGGSYY211 pKa = 11.02SKK213 pKa = 10.18FTVQAGLEE221 pKa = 4.06VRR223 pKa = 11.84GGYY226 pKa = 8.54GQNFQRR232 pKa = 11.84GVLATSPSIATVTAAFDD249 pKa = 3.5ATVNGPVGVIADD261 pKa = 5.29GISTATRR268 pKa = 11.84VADD271 pKa = 3.92LSIVGTTASAGSSSYY286 pKa = 11.38GVFVRR291 pKa = 11.84NSTNALVLDD300 pKa = 4.18SLNIAGGVAGAGSAGAAGSSGWSGTAGTGSNGGDD334 pKa = 3.2GFEE337 pKa = 5.28PGGACNTSSAGGGGAGGGGANSGGSGGKK365 pKa = 10.24GGVVDD370 pKa = 6.53GSCGWTGLCSNCDD383 pKa = 3.42AQTGNSGGSGGGVGGGGGGAAGSAATDD410 pKa = 3.79GLVGICTLGGTGPTLRR426 pKa = 11.84GGSGSSGANGTAGGAGSGGSAGANGATGLLGANGAGGGAGGGGGANDD473 pKa = 4.97CNIDD477 pKa = 3.63DD478 pKa = 4.77AGAGGGGGGGGGTRR492 pKa = 11.84ATAGGAGGGAGAQSIGLRR510 pKa = 11.84LQNASPTLIAIQIQLGTGGAGGTGGSGAAGQPGGSGGSGGVAFDD554 pKa = 3.9RR555 pKa = 11.84GGGGGNGGTGGTGGASGAGGGGGGGAAIGLSRR587 pKa = 11.84DD588 pKa = 3.52GSSTPSGSPSYY599 pKa = 10.73SGGVGGSGGSAGSGGNPGATGLVTNTTVAA628 pKa = 3.31
Molecular weight: 55.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0J0V049|A0A0J0V049_9ACTN Homoserine dehydrogenase OS=Actinobacteria bacterium IMCC26207 OX=1641811 GN=IMCC26207_10385 PE=3 SV=1
MM1 pKa = 7.45QPAQRR6 pKa = 11.84LASSKK11 pKa = 10.58SLIWLRR17 pKa = 11.84LVGLAPGRR25 pKa = 11.84VSRR28 pKa = 11.84AAILLGLGAFIAAVYY43 pKa = 10.27LPGSVVPDD51 pKa = 3.72TLDD54 pKa = 2.98MCGQAVSGVYY64 pKa = 10.35NDD66 pKa = 3.22WHH68 pKa = 6.85SPAIAGLWGFFNPRR82 pKa = 11.84IEE84 pKa = 4.7VIFLTTLAATILAVHH99 pKa = 7.19LILTRR104 pKa = 11.84WLRR107 pKa = 11.84PWVAVACTAAIMLFPATVGWMGHH130 pKa = 4.62VGKK133 pKa = 10.44DD134 pKa = 2.99EE135 pKa = 3.91WFAAAFLLGTALLARR150 pKa = 11.84AGTEE154 pKa = 3.33RR155 pKa = 11.84RR156 pKa = 11.84VRR158 pKa = 11.84LRR160 pKa = 11.84RR161 pKa = 11.84ALLMGAMICFWFAIAARR178 pKa = 11.84KK179 pKa = 8.68NALLPVGAALLVAWPVPGSVFGRR202 pKa = 11.84LQDD205 pKa = 3.34RR206 pKa = 11.84LLVRR210 pKa = 11.84RR211 pKa = 11.84VLAAGGVLILLVGSVSLFTSAIVRR235 pKa = 11.84PRR237 pKa = 11.84PRR239 pKa = 11.84HH240 pKa = 5.67PEE242 pKa = 3.55QQAFLFDD249 pKa = 3.81LTGLALAQQQMLFPPGVFPDD269 pKa = 3.65GTTLSGIGQSFDD281 pKa = 3.61PKK283 pKa = 10.05TGDD286 pKa = 3.21VFFFAPGTPANPFMLAEE303 pKa = 4.0QVSAVKK309 pKa = 10.08EE310 pKa = 3.97KK311 pKa = 10.16WLEE314 pKa = 3.83AVLAHH319 pKa = 7.01PDD321 pKa = 3.0SYY323 pKa = 11.65LRR325 pKa = 11.84TRR327 pKa = 11.84LSYY330 pKa = 10.52TSALLGIAGAHH341 pKa = 6.5PYY343 pKa = 9.69WSINDD348 pKa = 4.17PGSLPATWGSNCRR361 pKa = 11.84VPNRR365 pKa = 11.84TFPSLHH371 pKa = 5.71QRR373 pKa = 11.84VLDD376 pKa = 3.68LLLRR380 pKa = 11.84VEE382 pKa = 4.53RR383 pKa = 11.84GNLFRR388 pKa = 11.84GWTFLVILIAGSLLAGVRR406 pKa = 11.84KK407 pKa = 8.48VTEE410 pKa = 3.9ARR412 pKa = 11.84VLLLGGLFSFAGIAIAGISPTFRR435 pKa = 11.84YY436 pKa = 8.95SWFMAVCALIVAALALRR453 pKa = 11.84RR454 pKa = 11.84IPFAARR460 pKa = 11.84PDD462 pKa = 3.93PPGEE466 pKa = 4.11PLASVPDD473 pKa = 3.97SDD475 pKa = 5.48ASRR478 pKa = 11.84SDD480 pKa = 3.64PSPSSFSATMPTGEE494 pKa = 4.48PRR496 pKa = 11.84SRR498 pKa = 11.84WEE500 pKa = 3.78KK501 pKa = 10.85VLVQMGLIPDD511 pKa = 3.89SS512 pKa = 3.78
MM1 pKa = 7.45QPAQRR6 pKa = 11.84LASSKK11 pKa = 10.58SLIWLRR17 pKa = 11.84LVGLAPGRR25 pKa = 11.84VSRR28 pKa = 11.84AAILLGLGAFIAAVYY43 pKa = 10.27LPGSVVPDD51 pKa = 3.72TLDD54 pKa = 2.98MCGQAVSGVYY64 pKa = 10.35NDD66 pKa = 3.22WHH68 pKa = 6.85SPAIAGLWGFFNPRR82 pKa = 11.84IEE84 pKa = 4.7VIFLTTLAATILAVHH99 pKa = 7.19LILTRR104 pKa = 11.84WLRR107 pKa = 11.84PWVAVACTAAIMLFPATVGWMGHH130 pKa = 4.62VGKK133 pKa = 10.44DD134 pKa = 2.99EE135 pKa = 3.91WFAAAFLLGTALLARR150 pKa = 11.84AGTEE154 pKa = 3.33RR155 pKa = 11.84RR156 pKa = 11.84VRR158 pKa = 11.84LRR160 pKa = 11.84RR161 pKa = 11.84ALLMGAMICFWFAIAARR178 pKa = 11.84KK179 pKa = 8.68NALLPVGAALLVAWPVPGSVFGRR202 pKa = 11.84LQDD205 pKa = 3.34RR206 pKa = 11.84LLVRR210 pKa = 11.84RR211 pKa = 11.84VLAAGGVLILLVGSVSLFTSAIVRR235 pKa = 11.84PRR237 pKa = 11.84PRR239 pKa = 11.84HH240 pKa = 5.67PEE242 pKa = 3.55QQAFLFDD249 pKa = 3.81LTGLALAQQQMLFPPGVFPDD269 pKa = 3.65GTTLSGIGQSFDD281 pKa = 3.61PKK283 pKa = 10.05TGDD286 pKa = 3.21VFFFAPGTPANPFMLAEE303 pKa = 4.0QVSAVKK309 pKa = 10.08EE310 pKa = 3.97KK311 pKa = 10.16WLEE314 pKa = 3.83AVLAHH319 pKa = 7.01PDD321 pKa = 3.0SYY323 pKa = 11.65LRR325 pKa = 11.84TRR327 pKa = 11.84LSYY330 pKa = 10.52TSALLGIAGAHH341 pKa = 6.5PYY343 pKa = 9.69WSINDD348 pKa = 4.17PGSLPATWGSNCRR361 pKa = 11.84VPNRR365 pKa = 11.84TFPSLHH371 pKa = 5.71QRR373 pKa = 11.84VLDD376 pKa = 3.68LLLRR380 pKa = 11.84VEE382 pKa = 4.53RR383 pKa = 11.84GNLFRR388 pKa = 11.84GWTFLVILIAGSLLAGVRR406 pKa = 11.84KK407 pKa = 8.48VTEE410 pKa = 3.9ARR412 pKa = 11.84VLLLGGLFSFAGIAIAGISPTFRR435 pKa = 11.84YY436 pKa = 8.95SWFMAVCALIVAALALRR453 pKa = 11.84RR454 pKa = 11.84IPFAARR460 pKa = 11.84PDD462 pKa = 3.93PPGEE466 pKa = 4.11PLASVPDD473 pKa = 3.97SDD475 pKa = 5.48ASRR478 pKa = 11.84SDD480 pKa = 3.64PSPSSFSATMPTGEE494 pKa = 4.48PRR496 pKa = 11.84SRR498 pKa = 11.84WEE500 pKa = 3.78KK501 pKa = 10.85VLVQMGLIPDD511 pKa = 3.89SS512 pKa = 3.78
Molecular weight: 55.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1027166 |
40 |
2413 |
345.3 |
37.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.81 ± 0.046 | 0.93 ± 0.013 |
5.63 ± 0.032 | 6.141 ± 0.043 |
3.294 ± 0.029 | 8.563 ± 0.049 |
1.908 ± 0.023 | 4.573 ± 0.032 |
2.344 ± 0.029 | 10.433 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.049 ± 0.021 | 2.543 ± 0.03 |
5.452 ± 0.031 | 3.716 ± 0.027 |
6.167 ± 0.041 | 6.909 ± 0.04 |
5.933 ± 0.051 | 8.205 ± 0.04 |
1.487 ± 0.02 | 1.912 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
