
Pedobacter sp. AR-3-17
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Pedobacter; unclassified Pedobacter
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
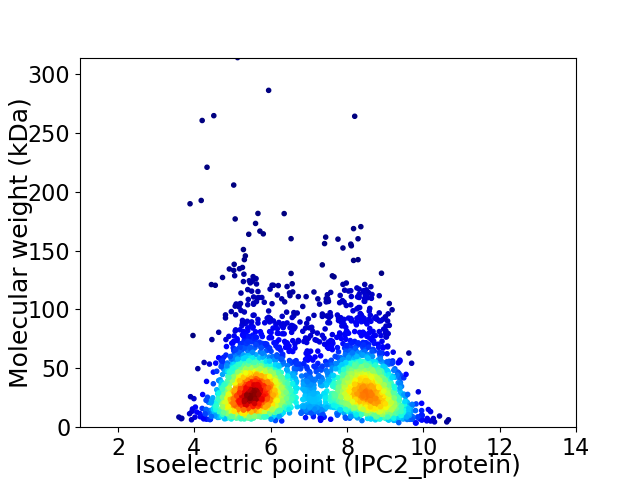
Virtual 2D-PAGE plot for 3357 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
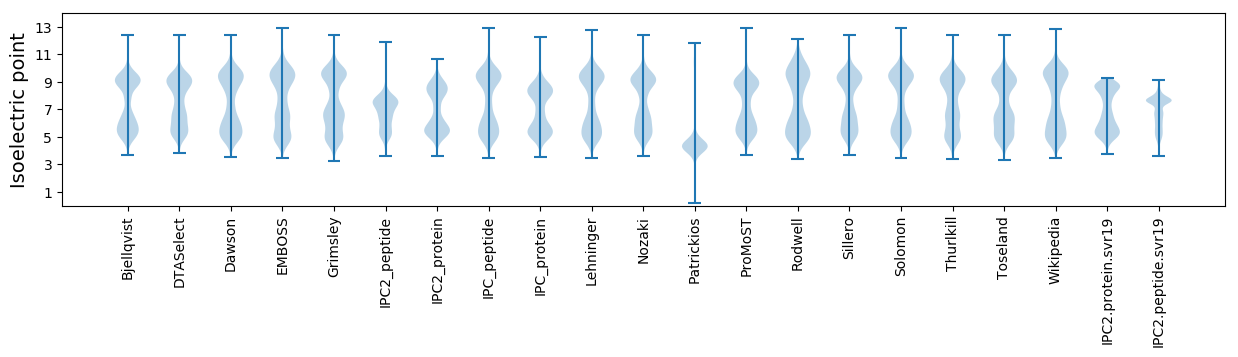
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U1BTU2|A0A4U1BTU2_9SPHI Uncharacterized protein OS=Pedobacter sp. AR-3-17 OX=2571271 GN=FA046_17095 PE=4 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84KK3 pKa = 9.21IFSTFIMIICLGVSVQSFAQTCNTVPVINSFSPNTGFIGSKK44 pKa = 10.28VSIFGANFDD53 pKa = 4.87PNTISNNVVYY63 pKa = 10.36FGSTKK68 pKa = 9.88ATVVSATFGQLEE80 pKa = 4.53VIVPVGASTAPISVTNQCNRR100 pKa = 11.84TAYY103 pKa = 9.47STVSFNGIFCPTPLDD118 pKa = 3.43NQTYY122 pKa = 9.64NNRR125 pKa = 11.84SFDD128 pKa = 3.65LQGISGAYY136 pKa = 10.53NMISQDD142 pKa = 3.78LDD144 pKa = 3.99LDD146 pKa = 4.02GKK148 pKa = 10.2PEE150 pKa = 4.28VISSNGGSLTVARR163 pKa = 11.84NNSTPGALSFTALNTNGGGAGIYY186 pKa = 10.34AADD189 pKa = 4.04FDD191 pKa = 5.23GDD193 pKa = 3.9GKK195 pKa = 11.17KK196 pKa = 10.62DD197 pKa = 3.34ILATSQVSRR206 pKa = 11.84NTSTLGNISLATPQSIPGVSGYY228 pKa = 10.53QIAAGDD234 pKa = 3.85FNNDD238 pKa = 2.55GKK240 pKa = 10.7IDD242 pKa = 4.03IIGEE246 pKa = 3.93SGGFVRR252 pKa = 11.84VAFNTSTGPGNISFGPAQLFGNGSVTFRR280 pKa = 11.84CTGIQVADD288 pKa = 3.39VDD290 pKa = 4.55GDD292 pKa = 3.87GKK294 pKa = 9.47TDD296 pKa = 4.04FLASQGDD303 pKa = 3.86GNRR306 pKa = 11.84AVTIRR311 pKa = 11.84NTTTVGSFTPSAEE324 pKa = 3.99APEE327 pKa = 4.86FWASDD332 pKa = 3.42SNAADD337 pKa = 3.61GFGTYY342 pKa = 9.78PYY344 pKa = 9.77RR345 pKa = 11.84SQIADD350 pKa = 3.3FDD352 pKa = 4.48KK353 pKa = 11.06DD354 pKa = 3.5GKK356 pKa = 10.57IDD358 pKa = 3.85FTSCNYY364 pKa = 10.92SGDD367 pKa = 3.71TNVAIWRR374 pKa = 11.84NISTVGNITFAPTVNFPSPINNYY397 pKa = 9.87RR398 pKa = 11.84IGVGDD403 pKa = 3.71VDD405 pKa = 3.85GDD407 pKa = 4.11GYY409 pKa = 10.25PDD411 pKa = 3.72VVTKK415 pKa = 10.79SLGVNVFSVYY425 pKa = 10.95KK426 pKa = 9.19NTTSAVGAPTFATRR440 pKa = 11.84IDD442 pKa = 4.14YY443 pKa = 8.43TSSWQAEE450 pKa = 4.02VSGIVIGDD458 pKa = 3.81LDD460 pKa = 4.43GDD462 pKa = 4.44FVPDD466 pKa = 3.31IATSGTNSSRR476 pKa = 11.84ILFHH480 pKa = 7.4RR481 pKa = 11.84NTSFQADD488 pKa = 3.57NTAPTAIAQNVVAALAPNGTVTVTAAQVDD517 pKa = 4.1NGSSDD522 pKa = 3.22ACGIKK527 pKa = 9.78TITLSQTVFTCANIGANPVTLTVTDD552 pKa = 3.4NAGNVTTANAIVNVQPAAIIVSGQTTVCQGGTIPLIANLGDD593 pKa = 3.59TYY595 pKa = 11.17QWYY598 pKa = 10.15NNGVAISGATNQNYY612 pKa = 7.48TATQTGSYY620 pKa = 8.47TVAVTNSGGCSGTSLPTVVTVNDD643 pKa = 3.65NPVVEE648 pKa = 4.04ITGRR652 pKa = 11.84VDD654 pKa = 3.84AYY656 pKa = 11.19CPGEE660 pKa = 4.05ALIASQSSLYY670 pKa = 9.59QWSKK674 pKa = 10.83NGVIVANATLRR685 pKa = 11.84EE686 pKa = 4.53FIPNGPGTYY695 pKa = 9.7SVAVIDD701 pKa = 5.53LFGCSASGSVTISPDD716 pKa = 3.16TTIPTIALNGAAQLTVGSDD735 pKa = 3.38TQFNDD740 pKa = 3.54PSVTANDD747 pKa = 3.53NCSTNIVVTSNLNLSTPGVYY767 pKa = 9.72TISYY771 pKa = 9.08KK772 pKa = 10.88AVDD775 pKa = 3.72GSGNEE780 pKa = 4.13SVTITRR786 pKa = 11.84QVTVLDD792 pKa = 3.5ATAPIVNAGRR802 pKa = 11.84VTVQLDD808 pKa = 3.31ANGQVTVVPASLNNGSIDD826 pKa = 3.65NSNGALTFSLDD837 pKa = 3.63KK838 pKa = 11.29SVFDD842 pKa = 6.04CIDD845 pKa = 3.78LNSGTGGTLTAITSVGTVIRR865 pKa = 11.84EE866 pKa = 4.13NYY868 pKa = 9.84SVGAGFNPNTGEE880 pKa = 3.55YY881 pKa = 9.73WMPQWSGSTVYY892 pKa = 11.03KK893 pKa = 10.58LDD895 pKa = 3.23VAGNVLGSFNSGQNQMMQLWMDD917 pKa = 4.2ADD919 pKa = 4.11SEE921 pKa = 4.52TDD923 pKa = 3.6YY924 pKa = 11.0YY925 pKa = 8.77TANWTSNNPYY935 pKa = 10.1NFTRR939 pKa = 11.84INEE942 pKa = 4.18NGQKK946 pKa = 9.55VWTYY950 pKa = 11.06NVANGQYY957 pKa = 10.73ASAISTDD964 pKa = 3.04ANFAYY969 pKa = 10.73VIGYY973 pKa = 8.46GGNRR977 pKa = 11.84IDD979 pKa = 4.74VLNKK983 pKa = 7.99TTGQFQRR990 pKa = 11.84SISLPGQIYY999 pKa = 10.17LYY1001 pKa = 10.77GSLVVANDD1009 pKa = 4.19HH1010 pKa = 7.03IYY1012 pKa = 10.82FGGQTNGWGNVPNTWNAIHH1031 pKa = 6.53EE1032 pKa = 4.38FKK1034 pKa = 10.5IDD1036 pKa = 3.52GTYY1039 pKa = 10.38VGSISTPQAVYY1050 pKa = 10.13NASFDD1055 pKa = 4.32GEE1057 pKa = 4.72VIWASAFSNQNYY1069 pKa = 9.59GYY1071 pKa = 10.36KK1072 pKa = 10.03ISDD1075 pKa = 3.4GNAYY1079 pKa = 10.3SGNKK1083 pKa = 9.18GIEE1086 pKa = 4.29VVLTATDD1093 pKa = 3.43AAGNSATDD1101 pKa = 3.14VGTVVVEE1108 pKa = 4.62DD1109 pKa = 3.98NLAPTITLNGNQVVQVVVGATFTDD1133 pKa = 4.13PGANAIDD1140 pKa = 3.83NCSATVEE1147 pKa = 4.39TSGSVNVNLAGTYY1160 pKa = 8.91TILYY1164 pKa = 9.05KK1165 pKa = 10.84AIDD1168 pKa = 3.89GSGNEE1173 pKa = 4.01SAQISRR1179 pKa = 11.84TVQVIEE1185 pKa = 5.28LDD1187 pKa = 3.8VKK1189 pKa = 9.84TKK1191 pKa = 10.85NITVKK1196 pKa = 10.65LDD1198 pKa = 3.5ANGSYY1203 pKa = 9.27TLDD1206 pKa = 3.65PSEE1209 pKa = 4.35VDD1211 pKa = 2.97NGSILGGGSLSLNITQLDD1229 pKa = 4.0CSNIGNPVTVTVTATSSSGNTASGTAIVTVEE1260 pKa = 4.65DD1261 pKa = 3.44KK1262 pKa = 10.64FAPIVVTQNVTIQLNAEE1279 pKa = 4.26GFGSISPSSINNGSSDD1295 pKa = 3.38NCGIATIGLSKK1306 pKa = 9.65TAFDD1310 pKa = 4.68CSNIGTNTVTLTVIDD1325 pKa = 3.83VNGNVSTATAVVTVEE1340 pKa = 4.43DD1341 pKa = 3.83KK1342 pKa = 11.04VVPIVVTQNRR1352 pKa = 11.84TIQLDD1357 pKa = 3.5AAGNASITAAAINNGSSDD1375 pKa = 3.24NCAIASVVLDD1385 pKa = 3.67KK1386 pKa = 11.38TSFTCEE1392 pKa = 3.65NIGANTVTLTVTDD1405 pKa = 3.47VNGNVSSATAVVTVEE1420 pKa = 5.01DD1421 pKa = 4.55KK1422 pKa = 10.67IAPIVITQNRR1432 pKa = 11.84TIQLDD1437 pKa = 3.5AAGNASITAAAINNGSSDD1455 pKa = 3.32NCAIATVVLDD1465 pKa = 3.64QTSFTCEE1472 pKa = 3.41QVGANTVTLTVTDD1485 pKa = 3.47VNGNVSSATAVVTVEE1500 pKa = 5.01DD1501 pKa = 4.55KK1502 pKa = 10.67IAPIVITQNRR1512 pKa = 11.84TIQLDD1517 pKa = 3.5AAGNASITAAAINNGSSDD1535 pKa = 3.32NCAIATVVLDD1545 pKa = 3.64QTSFTCEE1552 pKa = 3.54QVGVNTVTLTVTDD1565 pKa = 3.47VNGNVSSATAVVTVEE1580 pKa = 5.01DD1581 pKa = 4.55KK1582 pKa = 10.67IAPIVITQNRR1592 pKa = 11.84TIQLDD1597 pKa = 3.5AAGNASITAAAINNGSSDD1615 pKa = 3.32NCAIATVVLDD1625 pKa = 3.64QTSFTCEE1632 pKa = 3.54QVGVNTVTLTVTDD1645 pKa = 3.53VNGNVSTATAVVTVEE1660 pKa = 5.17DD1661 pKa = 4.27NIKK1664 pKa = 10.49PIVITQNATIQLDD1677 pKa = 3.63AAGNASITAVAINNGSTDD1695 pKa = 2.93NCAIATIVLSKK1706 pKa = 9.74TAFDD1710 pKa = 4.37CSNVGANTVTLTVTDD1725 pKa = 3.53VNGNVSTATAVVTVVDD1741 pKa = 5.76AIAPIITQLAFQEE1754 pKa = 4.33ICFVASNTYY1763 pKa = 10.64SLPTLTATDD1772 pKa = 3.66NCSVASVNYY1781 pKa = 8.75VLSGATSRR1789 pKa = 11.84IGTGTDD1795 pKa = 2.97ASGTLNTGLTTISWTVTDD1813 pKa = 3.46VNGNVSTSSNDD1824 pKa = 2.75IRR1826 pKa = 11.84IWPLPVLSITPSNADD1841 pKa = 2.95AFF1843 pKa = 4.26
MM1 pKa = 7.71RR2 pKa = 11.84KK3 pKa = 9.21IFSTFIMIICLGVSVQSFAQTCNTVPVINSFSPNTGFIGSKK44 pKa = 10.28VSIFGANFDD53 pKa = 4.87PNTISNNVVYY63 pKa = 10.36FGSTKK68 pKa = 9.88ATVVSATFGQLEE80 pKa = 4.53VIVPVGASTAPISVTNQCNRR100 pKa = 11.84TAYY103 pKa = 9.47STVSFNGIFCPTPLDD118 pKa = 3.43NQTYY122 pKa = 9.64NNRR125 pKa = 11.84SFDD128 pKa = 3.65LQGISGAYY136 pKa = 10.53NMISQDD142 pKa = 3.78LDD144 pKa = 3.99LDD146 pKa = 4.02GKK148 pKa = 10.2PEE150 pKa = 4.28VISSNGGSLTVARR163 pKa = 11.84NNSTPGALSFTALNTNGGGAGIYY186 pKa = 10.34AADD189 pKa = 4.04FDD191 pKa = 5.23GDD193 pKa = 3.9GKK195 pKa = 11.17KK196 pKa = 10.62DD197 pKa = 3.34ILATSQVSRR206 pKa = 11.84NTSTLGNISLATPQSIPGVSGYY228 pKa = 10.53QIAAGDD234 pKa = 3.85FNNDD238 pKa = 2.55GKK240 pKa = 10.7IDD242 pKa = 4.03IIGEE246 pKa = 3.93SGGFVRR252 pKa = 11.84VAFNTSTGPGNISFGPAQLFGNGSVTFRR280 pKa = 11.84CTGIQVADD288 pKa = 3.39VDD290 pKa = 4.55GDD292 pKa = 3.87GKK294 pKa = 9.47TDD296 pKa = 4.04FLASQGDD303 pKa = 3.86GNRR306 pKa = 11.84AVTIRR311 pKa = 11.84NTTTVGSFTPSAEE324 pKa = 3.99APEE327 pKa = 4.86FWASDD332 pKa = 3.42SNAADD337 pKa = 3.61GFGTYY342 pKa = 9.78PYY344 pKa = 9.77RR345 pKa = 11.84SQIADD350 pKa = 3.3FDD352 pKa = 4.48KK353 pKa = 11.06DD354 pKa = 3.5GKK356 pKa = 10.57IDD358 pKa = 3.85FTSCNYY364 pKa = 10.92SGDD367 pKa = 3.71TNVAIWRR374 pKa = 11.84NISTVGNITFAPTVNFPSPINNYY397 pKa = 9.87RR398 pKa = 11.84IGVGDD403 pKa = 3.71VDD405 pKa = 3.85GDD407 pKa = 4.11GYY409 pKa = 10.25PDD411 pKa = 3.72VVTKK415 pKa = 10.79SLGVNVFSVYY425 pKa = 10.95KK426 pKa = 9.19NTTSAVGAPTFATRR440 pKa = 11.84IDD442 pKa = 4.14YY443 pKa = 8.43TSSWQAEE450 pKa = 4.02VSGIVIGDD458 pKa = 3.81LDD460 pKa = 4.43GDD462 pKa = 4.44FVPDD466 pKa = 3.31IATSGTNSSRR476 pKa = 11.84ILFHH480 pKa = 7.4RR481 pKa = 11.84NTSFQADD488 pKa = 3.57NTAPTAIAQNVVAALAPNGTVTVTAAQVDD517 pKa = 4.1NGSSDD522 pKa = 3.22ACGIKK527 pKa = 9.78TITLSQTVFTCANIGANPVTLTVTDD552 pKa = 3.4NAGNVTTANAIVNVQPAAIIVSGQTTVCQGGTIPLIANLGDD593 pKa = 3.59TYY595 pKa = 11.17QWYY598 pKa = 10.15NNGVAISGATNQNYY612 pKa = 7.48TATQTGSYY620 pKa = 8.47TVAVTNSGGCSGTSLPTVVTVNDD643 pKa = 3.65NPVVEE648 pKa = 4.04ITGRR652 pKa = 11.84VDD654 pKa = 3.84AYY656 pKa = 11.19CPGEE660 pKa = 4.05ALIASQSSLYY670 pKa = 9.59QWSKK674 pKa = 10.83NGVIVANATLRR685 pKa = 11.84EE686 pKa = 4.53FIPNGPGTYY695 pKa = 9.7SVAVIDD701 pKa = 5.53LFGCSASGSVTISPDD716 pKa = 3.16TTIPTIALNGAAQLTVGSDD735 pKa = 3.38TQFNDD740 pKa = 3.54PSVTANDD747 pKa = 3.53NCSTNIVVTSNLNLSTPGVYY767 pKa = 9.72TISYY771 pKa = 9.08KK772 pKa = 10.88AVDD775 pKa = 3.72GSGNEE780 pKa = 4.13SVTITRR786 pKa = 11.84QVTVLDD792 pKa = 3.5ATAPIVNAGRR802 pKa = 11.84VTVQLDD808 pKa = 3.31ANGQVTVVPASLNNGSIDD826 pKa = 3.65NSNGALTFSLDD837 pKa = 3.63KK838 pKa = 11.29SVFDD842 pKa = 6.04CIDD845 pKa = 3.78LNSGTGGTLTAITSVGTVIRR865 pKa = 11.84EE866 pKa = 4.13NYY868 pKa = 9.84SVGAGFNPNTGEE880 pKa = 3.55YY881 pKa = 9.73WMPQWSGSTVYY892 pKa = 11.03KK893 pKa = 10.58LDD895 pKa = 3.23VAGNVLGSFNSGQNQMMQLWMDD917 pKa = 4.2ADD919 pKa = 4.11SEE921 pKa = 4.52TDD923 pKa = 3.6YY924 pKa = 11.0YY925 pKa = 8.77TANWTSNNPYY935 pKa = 10.1NFTRR939 pKa = 11.84INEE942 pKa = 4.18NGQKK946 pKa = 9.55VWTYY950 pKa = 11.06NVANGQYY957 pKa = 10.73ASAISTDD964 pKa = 3.04ANFAYY969 pKa = 10.73VIGYY973 pKa = 8.46GGNRR977 pKa = 11.84IDD979 pKa = 4.74VLNKK983 pKa = 7.99TTGQFQRR990 pKa = 11.84SISLPGQIYY999 pKa = 10.17LYY1001 pKa = 10.77GSLVVANDD1009 pKa = 4.19HH1010 pKa = 7.03IYY1012 pKa = 10.82FGGQTNGWGNVPNTWNAIHH1031 pKa = 6.53EE1032 pKa = 4.38FKK1034 pKa = 10.5IDD1036 pKa = 3.52GTYY1039 pKa = 10.38VGSISTPQAVYY1050 pKa = 10.13NASFDD1055 pKa = 4.32GEE1057 pKa = 4.72VIWASAFSNQNYY1069 pKa = 9.59GYY1071 pKa = 10.36KK1072 pKa = 10.03ISDD1075 pKa = 3.4GNAYY1079 pKa = 10.3SGNKK1083 pKa = 9.18GIEE1086 pKa = 4.29VVLTATDD1093 pKa = 3.43AAGNSATDD1101 pKa = 3.14VGTVVVEE1108 pKa = 4.62DD1109 pKa = 3.98NLAPTITLNGNQVVQVVVGATFTDD1133 pKa = 4.13PGANAIDD1140 pKa = 3.83NCSATVEE1147 pKa = 4.39TSGSVNVNLAGTYY1160 pKa = 8.91TILYY1164 pKa = 9.05KK1165 pKa = 10.84AIDD1168 pKa = 3.89GSGNEE1173 pKa = 4.01SAQISRR1179 pKa = 11.84TVQVIEE1185 pKa = 5.28LDD1187 pKa = 3.8VKK1189 pKa = 9.84TKK1191 pKa = 10.85NITVKK1196 pKa = 10.65LDD1198 pKa = 3.5ANGSYY1203 pKa = 9.27TLDD1206 pKa = 3.65PSEE1209 pKa = 4.35VDD1211 pKa = 2.97NGSILGGGSLSLNITQLDD1229 pKa = 4.0CSNIGNPVTVTVTATSSSGNTASGTAIVTVEE1260 pKa = 4.65DD1261 pKa = 3.44KK1262 pKa = 10.64FAPIVVTQNVTIQLNAEE1279 pKa = 4.26GFGSISPSSINNGSSDD1295 pKa = 3.38NCGIATIGLSKK1306 pKa = 9.65TAFDD1310 pKa = 4.68CSNIGTNTVTLTVIDD1325 pKa = 3.83VNGNVSTATAVVTVEE1340 pKa = 4.43DD1341 pKa = 3.83KK1342 pKa = 11.04VVPIVVTQNRR1352 pKa = 11.84TIQLDD1357 pKa = 3.5AAGNASITAAAINNGSSDD1375 pKa = 3.24NCAIASVVLDD1385 pKa = 3.67KK1386 pKa = 11.38TSFTCEE1392 pKa = 3.65NIGANTVTLTVTDD1405 pKa = 3.47VNGNVSSATAVVTVEE1420 pKa = 5.01DD1421 pKa = 4.55KK1422 pKa = 10.67IAPIVITQNRR1432 pKa = 11.84TIQLDD1437 pKa = 3.5AAGNASITAAAINNGSSDD1455 pKa = 3.32NCAIATVVLDD1465 pKa = 3.64QTSFTCEE1472 pKa = 3.41QVGANTVTLTVTDD1485 pKa = 3.47VNGNVSSATAVVTVEE1500 pKa = 5.01DD1501 pKa = 4.55KK1502 pKa = 10.67IAPIVITQNRR1512 pKa = 11.84TIQLDD1517 pKa = 3.5AAGNASITAAAINNGSSDD1535 pKa = 3.32NCAIATVVLDD1545 pKa = 3.64QTSFTCEE1552 pKa = 3.54QVGVNTVTLTVTDD1565 pKa = 3.47VNGNVSSATAVVTVEE1580 pKa = 5.01DD1581 pKa = 4.55KK1582 pKa = 10.67IAPIVITQNRR1592 pKa = 11.84TIQLDD1597 pKa = 3.5AAGNASITAAAINNGSSDD1615 pKa = 3.32NCAIATVVLDD1625 pKa = 3.64QTSFTCEE1632 pKa = 3.54QVGVNTVTLTVTDD1645 pKa = 3.53VNGNVSTATAVVTVEE1660 pKa = 5.17DD1661 pKa = 4.27NIKK1664 pKa = 10.49PIVITQNATIQLDD1677 pKa = 3.63AAGNASITAVAINNGSTDD1695 pKa = 2.93NCAIATIVLSKK1706 pKa = 9.74TAFDD1710 pKa = 4.37CSNVGANTVTLTVTDD1725 pKa = 3.53VNGNVSTATAVVTVVDD1741 pKa = 5.76AIAPIITQLAFQEE1754 pKa = 4.33ICFVASNTYY1763 pKa = 10.64SLPTLTATDD1772 pKa = 3.66NCSVASVNYY1781 pKa = 8.75VLSGATSRR1789 pKa = 11.84IGTGTDD1795 pKa = 2.97ASGTLNTGLTTISWTVTDD1813 pKa = 3.46VNGNVSTSSNDD1824 pKa = 2.75IRR1826 pKa = 11.84IWPLPVLSITPSNADD1841 pKa = 2.95AFF1843 pKa = 4.26
Molecular weight: 189.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U1C526|A0A4U1C526_9SPHI Translation initiation factor OS=Pedobacter sp. AR-3-17 OX=2571271 GN=FA046_04900 PE=3 SV=1
MM1 pKa = 7.57AHH3 pKa = 7.64PKK5 pKa = 10.37RR6 pKa = 11.84KK7 pKa = 8.88ISKK10 pKa = 8.89SRR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.27KK15 pKa = 11.24RR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 6.02YY20 pKa = 10.71KK21 pKa = 10.15AVAPSLWTCKK31 pKa = 8.07EE32 pKa = 3.85TGAVHH37 pKa = 6.83LPHH40 pKa = 7.23RR41 pKa = 11.84AYY43 pKa = 10.88NVDD46 pKa = 2.83GNLYY50 pKa = 10.45FRR52 pKa = 11.84GKK54 pKa = 10.67LIIEE58 pKa = 4.3NTSIAA63 pKa = 3.65
MM1 pKa = 7.57AHH3 pKa = 7.64PKK5 pKa = 10.37RR6 pKa = 11.84KK7 pKa = 8.88ISKK10 pKa = 8.89SRR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.27KK15 pKa = 11.24RR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 6.02YY20 pKa = 10.71KK21 pKa = 10.15AVAPSLWTCKK31 pKa = 8.07EE32 pKa = 3.85TGAVHH37 pKa = 6.83LPHH40 pKa = 7.23RR41 pKa = 11.84AYY43 pKa = 10.88NVDD46 pKa = 2.83GNLYY50 pKa = 10.45FRR52 pKa = 11.84GKK54 pKa = 10.67LIIEE58 pKa = 4.3NTSIAA63 pKa = 3.65
Molecular weight: 7.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1173386 |
32 |
2876 |
349.5 |
39.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.956 ± 0.053 | 0.757 ± 0.014 |
5.275 ± 0.03 | 5.936 ± 0.046 |
5.272 ± 0.039 | 6.547 ± 0.048 |
1.683 ± 0.025 | 7.99 ± 0.038 |
7.936 ± 0.046 | 9.527 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.104 ± 0.024 | 6.251 ± 0.05 |
3.488 ± 0.024 | 3.674 ± 0.022 |
3.261 ± 0.028 | 6.526 ± 0.039 |
5.665 ± 0.05 | 6.087 ± 0.033 |
1.086 ± 0.016 | 3.981 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
