
Vibrio phage Saratov-12
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Zobellviridae; unclassified Zobellviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
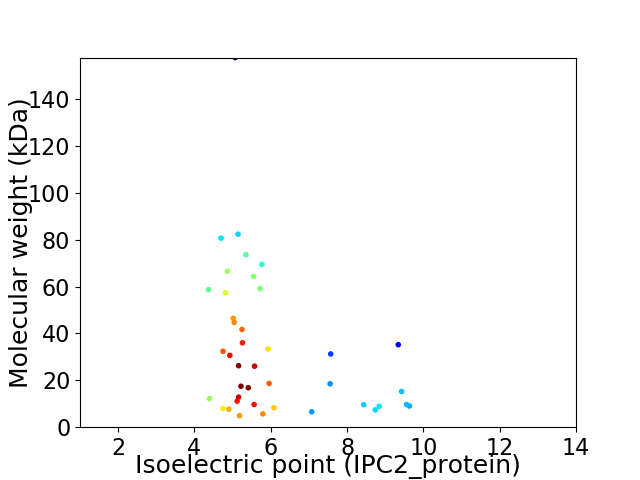
Virtual 2D-PAGE plot for 41 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XV87|A0A6G6XV87_9CAUD Uncharacterized protein OS=Vibrio phage Saratov-12 OX=2712943 GN=Saratov12_00067 PE=4 SV=1
MM1 pKa = 7.48AKK3 pKa = 9.63IVLSDD8 pKa = 3.52VTSGSSVTINRR19 pKa = 11.84NFQKK23 pKa = 10.4IEE25 pKa = 3.87DD26 pKa = 4.05ALNDD30 pKa = 3.53EE31 pKa = 4.7VLFRR35 pKa = 11.84DD36 pKa = 3.91NPEE39 pKa = 4.08GEE41 pKa = 4.29PNQMEE46 pKa = 4.67SDD48 pKa = 3.68LDD50 pKa = 3.88MNSNDD55 pKa = 3.37ILNAGVIKK63 pKa = 10.38AKK65 pKa = 10.34RR66 pKa = 11.84VIADD70 pKa = 3.63SIDD73 pKa = 3.47NNTEE77 pKa = 3.74SLIDD81 pKa = 3.89GGVSDD86 pKa = 4.72NEE88 pKa = 4.03IVKK91 pKa = 9.97WDD93 pKa = 3.29VDD95 pKa = 3.49TSKK98 pKa = 10.88FVGTGVFSNSEE109 pKa = 4.09GEE111 pKa = 4.4LDD113 pKa = 3.61VSPNSVSVGVHH124 pKa = 6.6KK125 pKa = 10.43IGSMGEE131 pKa = 3.8NVGFFNKK138 pKa = 9.09ATGKK142 pKa = 10.49SYY144 pKa = 10.17FPSWQGVGISDD155 pKa = 3.88SDD157 pKa = 3.73TFTAVTRR164 pKa = 11.84VHH166 pKa = 6.83GPLEE170 pKa = 4.23YY171 pKa = 10.39VVRR174 pKa = 11.84TTNVSEE180 pKa = 4.06VLINPEE186 pKa = 4.21FIVTVPYY193 pKa = 10.7SEE195 pKa = 4.43TVFKK199 pKa = 10.8IGLYY203 pKa = 8.39CTNEE207 pKa = 3.79VPNVHH212 pKa = 6.86FRR214 pKa = 11.84LTYY217 pKa = 9.68NNSIVFDD224 pKa = 4.03GDD226 pKa = 3.33MGSSQSVGMTTFTLPTPVDD245 pKa = 3.78LRR247 pKa = 11.84QGAVLVAKK255 pKa = 10.3AYY257 pKa = 10.52GIGQDD262 pKa = 4.11LYY264 pKa = 11.79VKK266 pKa = 10.26GGSDD270 pKa = 3.73GVPAYY275 pKa = 10.04SLWMRR280 pKa = 11.84PWVDD284 pKa = 3.24KK285 pKa = 10.99EE286 pKa = 4.04LATVEE291 pKa = 4.24YY292 pKa = 11.03VDD294 pKa = 5.28GKK296 pKa = 9.69PQGKK300 pKa = 7.68EE301 pKa = 3.9TFVEE305 pKa = 5.4LLDD308 pKa = 4.71CPNTYY313 pKa = 10.24VGQGKK318 pKa = 9.76KK319 pKa = 9.86LVAVNEE325 pKa = 4.38SEE327 pKa = 4.38SALEE331 pKa = 4.57FIPHH335 pKa = 5.63TLEE338 pKa = 4.26SLDD341 pKa = 3.44NVLIVSPIQTDD352 pKa = 4.35GEE354 pKa = 4.32VLMYY358 pKa = 11.08NLTEE362 pKa = 4.28NRR364 pKa = 11.84WEE366 pKa = 4.07NQALPEE372 pKa = 3.97FQEE375 pKa = 4.4DD376 pKa = 4.22IIRR379 pKa = 11.84ISYY382 pKa = 6.69TTPNSLPAKK391 pKa = 9.85QLLSTSNVGSVITFNVTEE409 pKa = 4.05IASDD413 pKa = 4.34FVQTDD418 pKa = 3.6LTSGVLTFVDD428 pKa = 3.5SAKK431 pKa = 11.25AMFTMSVQVIRR442 pKa = 11.84EE443 pKa = 3.95TGGAGDD449 pKa = 5.05ALWGMYY455 pKa = 10.2VEE457 pKa = 4.74TSMDD461 pKa = 4.33GISWSPVSGTTRR473 pKa = 11.84LLLLTGGDD481 pKa = 3.66SGDD484 pKa = 3.53VKK486 pKa = 11.08LIDD489 pKa = 3.63YY490 pKa = 7.38TVPISVVAGEE500 pKa = 4.22KK501 pKa = 10.59VRR503 pKa = 11.84FKK505 pKa = 10.98QYY507 pKa = 10.65TSDD510 pKa = 3.2SSKK513 pKa = 10.89QIGIISSPQTIAGIPTNAGVTLGVYY538 pKa = 9.72RR539 pKa = 11.84VQHH542 pKa = 5.67
MM1 pKa = 7.48AKK3 pKa = 9.63IVLSDD8 pKa = 3.52VTSGSSVTINRR19 pKa = 11.84NFQKK23 pKa = 10.4IEE25 pKa = 3.87DD26 pKa = 4.05ALNDD30 pKa = 3.53EE31 pKa = 4.7VLFRR35 pKa = 11.84DD36 pKa = 3.91NPEE39 pKa = 4.08GEE41 pKa = 4.29PNQMEE46 pKa = 4.67SDD48 pKa = 3.68LDD50 pKa = 3.88MNSNDD55 pKa = 3.37ILNAGVIKK63 pKa = 10.38AKK65 pKa = 10.34RR66 pKa = 11.84VIADD70 pKa = 3.63SIDD73 pKa = 3.47NNTEE77 pKa = 3.74SLIDD81 pKa = 3.89GGVSDD86 pKa = 4.72NEE88 pKa = 4.03IVKK91 pKa = 9.97WDD93 pKa = 3.29VDD95 pKa = 3.49TSKK98 pKa = 10.88FVGTGVFSNSEE109 pKa = 4.09GEE111 pKa = 4.4LDD113 pKa = 3.61VSPNSVSVGVHH124 pKa = 6.6KK125 pKa = 10.43IGSMGEE131 pKa = 3.8NVGFFNKK138 pKa = 9.09ATGKK142 pKa = 10.49SYY144 pKa = 10.17FPSWQGVGISDD155 pKa = 3.88SDD157 pKa = 3.73TFTAVTRR164 pKa = 11.84VHH166 pKa = 6.83GPLEE170 pKa = 4.23YY171 pKa = 10.39VVRR174 pKa = 11.84TTNVSEE180 pKa = 4.06VLINPEE186 pKa = 4.21FIVTVPYY193 pKa = 10.7SEE195 pKa = 4.43TVFKK199 pKa = 10.8IGLYY203 pKa = 8.39CTNEE207 pKa = 3.79VPNVHH212 pKa = 6.86FRR214 pKa = 11.84LTYY217 pKa = 9.68NNSIVFDD224 pKa = 4.03GDD226 pKa = 3.33MGSSQSVGMTTFTLPTPVDD245 pKa = 3.78LRR247 pKa = 11.84QGAVLVAKK255 pKa = 10.3AYY257 pKa = 10.52GIGQDD262 pKa = 4.11LYY264 pKa = 11.79VKK266 pKa = 10.26GGSDD270 pKa = 3.73GVPAYY275 pKa = 10.04SLWMRR280 pKa = 11.84PWVDD284 pKa = 3.24KK285 pKa = 10.99EE286 pKa = 4.04LATVEE291 pKa = 4.24YY292 pKa = 11.03VDD294 pKa = 5.28GKK296 pKa = 9.69PQGKK300 pKa = 7.68EE301 pKa = 3.9TFVEE305 pKa = 5.4LLDD308 pKa = 4.71CPNTYY313 pKa = 10.24VGQGKK318 pKa = 9.76KK319 pKa = 9.86LVAVNEE325 pKa = 4.38SEE327 pKa = 4.38SALEE331 pKa = 4.57FIPHH335 pKa = 5.63TLEE338 pKa = 4.26SLDD341 pKa = 3.44NVLIVSPIQTDD352 pKa = 4.35GEE354 pKa = 4.32VLMYY358 pKa = 11.08NLTEE362 pKa = 4.28NRR364 pKa = 11.84WEE366 pKa = 4.07NQALPEE372 pKa = 3.97FQEE375 pKa = 4.4DD376 pKa = 4.22IIRR379 pKa = 11.84ISYY382 pKa = 6.69TTPNSLPAKK391 pKa = 9.85QLLSTSNVGSVITFNVTEE409 pKa = 4.05IASDD413 pKa = 4.34FVQTDD418 pKa = 3.6LTSGVLTFVDD428 pKa = 3.5SAKK431 pKa = 11.25AMFTMSVQVIRR442 pKa = 11.84EE443 pKa = 3.95TGGAGDD449 pKa = 5.05ALWGMYY455 pKa = 10.2VEE457 pKa = 4.74TSMDD461 pKa = 4.33GISWSPVSGTTRR473 pKa = 11.84LLLLTGGDD481 pKa = 3.66SGDD484 pKa = 3.53VKK486 pKa = 11.08LIDD489 pKa = 3.63YY490 pKa = 7.38TVPISVVAGEE500 pKa = 4.22KK501 pKa = 10.59VRR503 pKa = 11.84FKK505 pKa = 10.98QYY507 pKa = 10.65TSDD510 pKa = 3.2SSKK513 pKa = 10.89QIGIISSPQTIAGIPTNAGVTLGVYY538 pKa = 9.72RR539 pKa = 11.84VQHH542 pKa = 5.67
Molecular weight: 58.73 kDa
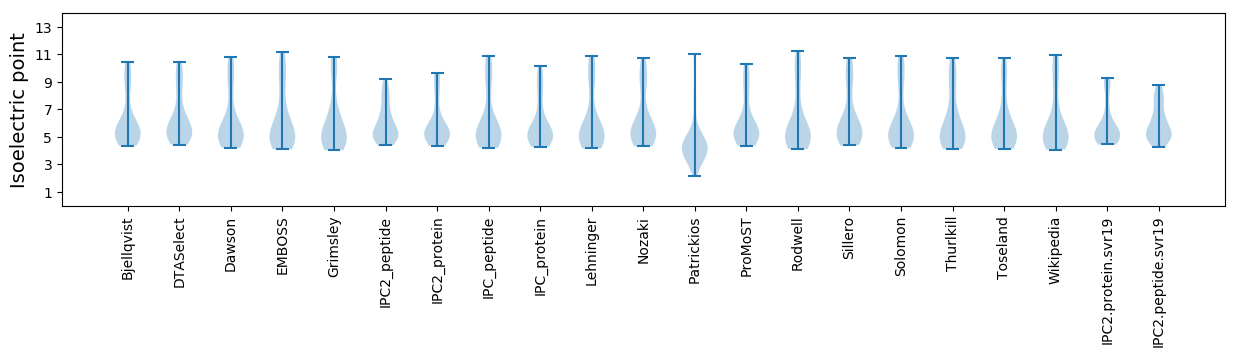
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XUJ4|A0A6G6XUJ4_9CAUD MazG domain-containing protein OS=Vibrio phage Saratov-12 OX=2712943 GN=Saratov12_00075 PE=4 SV=1
MM1 pKa = 7.82KK2 pKa = 10.23KK3 pKa = 10.13RR4 pKa = 11.84NPNRR8 pKa = 11.84AHH10 pKa = 5.68SPEE13 pKa = 3.78RR14 pKa = 11.84LMLRR18 pKa = 11.84KK19 pKa = 8.35QTADD23 pKa = 2.91VGLIKK28 pKa = 10.53VVNLISRR35 pKa = 11.84ATGGAVLGGRR45 pKa = 11.84LGFICAAIKK54 pKa = 10.24QDD56 pKa = 3.25GANIRR61 pKa = 11.84SLSTWEE67 pKa = 4.86AYY69 pKa = 9.18GQVYY73 pKa = 10.82DD74 pKa = 4.53SVTTMAIIHH83 pKa = 6.32
MM1 pKa = 7.82KK2 pKa = 10.23KK3 pKa = 10.13RR4 pKa = 11.84NPNRR8 pKa = 11.84AHH10 pKa = 5.68SPEE13 pKa = 3.78RR14 pKa = 11.84LMLRR18 pKa = 11.84KK19 pKa = 8.35QTADD23 pKa = 2.91VGLIKK28 pKa = 10.53VVNLISRR35 pKa = 11.84ATGGAVLGGRR45 pKa = 11.84LGFICAAIKK54 pKa = 10.24QDD56 pKa = 3.25GANIRR61 pKa = 11.84SLSTWEE67 pKa = 4.86AYY69 pKa = 9.18GQVYY73 pKa = 10.82DD74 pKa = 4.53SVTTMAIIHH83 pKa = 6.32
Molecular weight: 9.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12364 |
46 |
1416 |
301.6 |
33.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.225 ± 0.478 | 0.922 ± 0.227 |
6.398 ± 0.344 | 6.729 ± 0.367 |
3.664 ± 0.232 | 8.031 ± 0.767 |
1.432 ± 0.216 | 5.718 ± 0.209 |
6.082 ± 0.372 | 7.15 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.79 ± 0.279 | 4.926 ± 0.227 |
3.915 ± 0.383 | 4.44 ± 0.403 |
4.958 ± 0.277 | 6.317 ± 0.344 |
6.309 ± 0.426 | 6.956 ± 0.308 |
1.407 ± 0.109 | 3.632 ± 0.207 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
