
Ephemeroptericola cinctiostellae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Ephemeroptericola
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
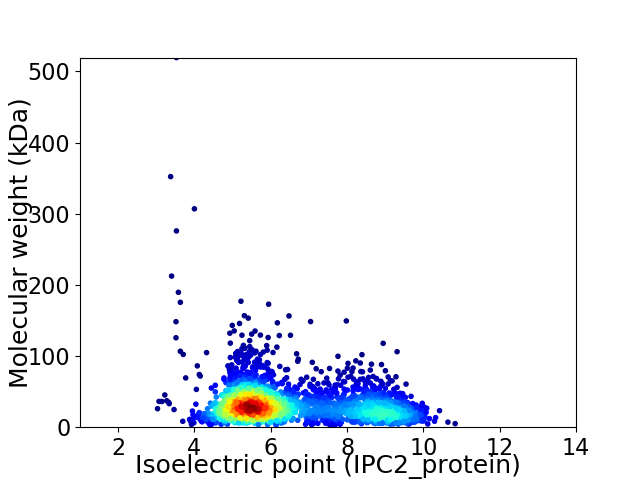
Virtual 2D-PAGE plot for 2476 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345DBY0|A0A345DBY0_9BURK Uncharacterized protein OS=Ephemeroptericola cinctiostellae OX=2268024 GN=DTO96_101608 PE=4 SV=1
MM1 pKa = 7.5IVLGTNNADD10 pKa = 3.75TLGSSGDD17 pKa = 3.53DD18 pKa = 3.45TLIGLAGDD26 pKa = 3.88DD27 pKa = 4.16TLNGYY32 pKa = 8.75LPWGTALGNTTYY44 pKa = 10.74RR45 pKa = 11.84FSKK48 pKa = 10.89GDD50 pKa = 3.33GNDD53 pKa = 3.3VVKK56 pKa = 10.8EE57 pKa = 4.02NAGSDD62 pKa = 3.64TIEE65 pKa = 4.06FTDD68 pKa = 3.64VKK70 pKa = 9.84STEE73 pKa = 4.06VFFKK77 pKa = 10.88LGNNGKK83 pKa = 9.52DD84 pKa = 3.54LIILYY89 pKa = 7.61GTNNSITISEE99 pKa = 4.23MIYY102 pKa = 9.31WPSGRR107 pKa = 11.84AIEE110 pKa = 4.15NFKK113 pKa = 11.07FSDD116 pKa = 3.94GVIWDD121 pKa = 3.95LNEE124 pKa = 4.71LYY126 pKa = 11.04NNTATTIGTLGNDD139 pKa = 4.76IISGWDD145 pKa = 3.34GFNVIDD151 pKa = 4.88GKK153 pKa = 11.24EE154 pKa = 4.07GDD156 pKa = 4.51DD157 pKa = 5.44LINGQTKK164 pKa = 10.6NDD166 pKa = 3.82TLLGGLGNDD175 pKa = 3.87TLYY178 pKa = 11.27GGYY181 pKa = 10.83GNDD184 pKa = 4.2ASDD187 pKa = 4.47GGDD190 pKa = 3.48GDD192 pKa = 5.37DD193 pKa = 4.18YY194 pKa = 11.64LYY196 pKa = 11.08DD197 pKa = 3.51SSGNNTLLGGAGNDD211 pKa = 3.53SLYY214 pKa = 11.23GGAGNDD220 pKa = 3.57VLNGGDD226 pKa = 3.82GNDD229 pKa = 3.44VLFGGDD235 pKa = 3.86GNDD238 pKa = 2.67IYY240 pKa = 11.12YY241 pKa = 10.2INDD244 pKa = 3.41IGDD247 pKa = 3.48QIYY250 pKa = 10.62DD251 pKa = 3.61SNGIADD257 pKa = 3.72TAYY260 pKa = 9.58ISVNGVRR267 pKa = 11.84TTGIEE272 pKa = 3.89NTVYY276 pKa = 10.88TNDD279 pKa = 3.93AQPLPYY285 pKa = 10.12FIRR288 pKa = 11.84ALDD291 pKa = 3.64SKK293 pKa = 7.9YY294 pKa = 8.08TWGAIGQGQTLNYY307 pKa = 10.32SFALNADD314 pKa = 3.41GGEE317 pKa = 4.31VGFEE321 pKa = 4.68LYY323 pKa = 10.14TEE325 pKa = 4.17EE326 pKa = 3.86QKK328 pKa = 9.8TAIRR332 pKa = 11.84EE333 pKa = 4.14ALQKK337 pKa = 10.37YY338 pKa = 8.63AQLVNLTFVEE348 pKa = 4.93VVDD351 pKa = 4.0STSVQLRR358 pKa = 11.84FFRR361 pKa = 11.84DD362 pKa = 3.56DD363 pKa = 3.85LSTAGYY369 pKa = 9.49AAYY372 pKa = 9.91AGYY375 pKa = 11.18ANYY378 pKa = 9.94PFDD381 pKa = 3.8SSKK384 pKa = 11.36GGDD387 pKa = 2.93IHH389 pKa = 7.72IKK391 pKa = 8.82NTISNLNQLDD401 pKa = 3.64GSFDD405 pKa = 3.74VLIHH409 pKa = 6.6EE410 pKa = 5.04IGHH413 pKa = 6.26ALGLKK418 pKa = 10.24HH419 pKa = 6.57PFEE422 pKa = 5.07DD423 pKa = 4.07TDD425 pKa = 3.91TLPTSEE431 pKa = 6.04DD432 pKa = 3.48STTYY436 pKa = 11.12SAMSYY441 pKa = 10.59NSLWQANKK449 pKa = 8.67NTVRR453 pKa = 11.84MYY455 pKa = 11.38DD456 pKa = 3.65LATLHH461 pKa = 6.09YY462 pKa = 9.38FYY464 pKa = 11.04GVNASQRR471 pKa = 11.84SEE473 pKa = 3.8NNTYY477 pKa = 11.08AFSDD481 pKa = 3.72ANFTDD486 pKa = 3.95HH487 pKa = 6.65YY488 pKa = 9.3VWDD491 pKa = 4.19GAGVDD496 pKa = 3.77VFDD499 pKa = 6.52ANSQTQNVSVNLNPGSWNFIGSKK522 pKa = 10.07AVSILSAGQSFIGFGTLIEE541 pKa = 4.25NAVSGSGNDD550 pKa = 3.67TVIGNALDD558 pKa = 3.97NQIHH562 pKa = 5.93TNAGNDD568 pKa = 3.49WLKK571 pKa = 11.29GGAGNDD577 pKa = 3.18SLYY580 pKa = 11.04AGEE583 pKa = 5.88GNDD586 pKa = 3.74TLNGQVGADD595 pKa = 3.42VMVGGNGDD603 pKa = 3.44DD604 pKa = 3.94YY605 pKa = 11.63YY606 pKa = 11.84YY607 pKa = 10.89FDD609 pKa = 3.9NVGDD613 pKa = 3.91AVIEE617 pKa = 4.23VVDD620 pKa = 4.23AGLDD624 pKa = 3.75CVEE627 pKa = 4.93ASANCTLNDD636 pKa = 3.79NIEE639 pKa = 4.02NLVLSGSGALSGMGNALNNQMKK661 pKa = 10.52GSAEE665 pKa = 4.0NNTLKK670 pKa = 10.82GMAGNDD676 pKa = 3.52SLLGFAGNDD685 pKa = 3.45ALDD688 pKa = 4.11GGTGNDD694 pKa = 3.63TLRR697 pKa = 11.84GGLGNDD703 pKa = 3.52VLNGGDD709 pKa = 3.82GIDD712 pKa = 3.28WADD715 pKa = 3.82YY716 pKa = 11.1VDD718 pKa = 3.89MTASVNVNLNITTVQVTGAGGSDD741 pKa = 3.08TLTNIEE747 pKa = 4.08NVRR750 pKa = 11.84AGSANDD756 pKa = 3.54NLKK759 pKa = 11.11GNASDD764 pKa = 4.61NVLDD768 pKa = 4.29GQAGNDD774 pKa = 3.48ALFGNAGNDD783 pKa = 3.55NLIGGAGNDD792 pKa = 3.6ILNGGLGNDD801 pKa = 3.86NLSGGDD807 pKa = 3.57GNDD810 pKa = 2.99WADD813 pKa = 3.78YY814 pKa = 10.22FDD816 pKa = 3.51STAAVSVDD824 pKa = 3.57LRR826 pKa = 11.84LATQQNTVGAGLDD839 pKa = 2.89IFYY842 pKa = 10.52NIEE845 pKa = 3.91NLRR848 pKa = 11.84GGLGNDD854 pKa = 3.6TLNGSVYY861 pKa = 11.12ANALVGQAGNDD872 pKa = 3.35FMVGNGGNDD881 pKa = 3.12TLYY884 pKa = 11.05AGLGNDD890 pKa = 3.71TLFGGTGNDD899 pKa = 2.84TYY901 pKa = 11.38QFNRR905 pKa = 11.84GDD907 pKa = 3.93GQDD910 pKa = 3.41TLTDD914 pKa = 3.4ADD916 pKa = 3.79ATVGNSDD923 pKa = 3.47TLAFQTGVTNSQLWLTHH940 pKa = 5.94TGNDD944 pKa = 3.35LVVSVIGTTDD954 pKa = 2.87KK955 pKa = 10.7VTIKK959 pKa = 10.26NWYY962 pKa = 8.22VSSANHH968 pKa = 5.79VEE970 pKa = 4.34SITAGGKK977 pKa = 7.53TLADD981 pKa = 3.73GNVNALVAAMSAFSPPAAGQTTLPSSYY1008 pKa = 8.48QTALNPVIAANWVV1021 pKa = 3.13
MM1 pKa = 7.5IVLGTNNADD10 pKa = 3.75TLGSSGDD17 pKa = 3.53DD18 pKa = 3.45TLIGLAGDD26 pKa = 3.88DD27 pKa = 4.16TLNGYY32 pKa = 8.75LPWGTALGNTTYY44 pKa = 10.74RR45 pKa = 11.84FSKK48 pKa = 10.89GDD50 pKa = 3.33GNDD53 pKa = 3.3VVKK56 pKa = 10.8EE57 pKa = 4.02NAGSDD62 pKa = 3.64TIEE65 pKa = 4.06FTDD68 pKa = 3.64VKK70 pKa = 9.84STEE73 pKa = 4.06VFFKK77 pKa = 10.88LGNNGKK83 pKa = 9.52DD84 pKa = 3.54LIILYY89 pKa = 7.61GTNNSITISEE99 pKa = 4.23MIYY102 pKa = 9.31WPSGRR107 pKa = 11.84AIEE110 pKa = 4.15NFKK113 pKa = 11.07FSDD116 pKa = 3.94GVIWDD121 pKa = 3.95LNEE124 pKa = 4.71LYY126 pKa = 11.04NNTATTIGTLGNDD139 pKa = 4.76IISGWDD145 pKa = 3.34GFNVIDD151 pKa = 4.88GKK153 pKa = 11.24EE154 pKa = 4.07GDD156 pKa = 4.51DD157 pKa = 5.44LINGQTKK164 pKa = 10.6NDD166 pKa = 3.82TLLGGLGNDD175 pKa = 3.87TLYY178 pKa = 11.27GGYY181 pKa = 10.83GNDD184 pKa = 4.2ASDD187 pKa = 4.47GGDD190 pKa = 3.48GDD192 pKa = 5.37DD193 pKa = 4.18YY194 pKa = 11.64LYY196 pKa = 11.08DD197 pKa = 3.51SSGNNTLLGGAGNDD211 pKa = 3.53SLYY214 pKa = 11.23GGAGNDD220 pKa = 3.57VLNGGDD226 pKa = 3.82GNDD229 pKa = 3.44VLFGGDD235 pKa = 3.86GNDD238 pKa = 2.67IYY240 pKa = 11.12YY241 pKa = 10.2INDD244 pKa = 3.41IGDD247 pKa = 3.48QIYY250 pKa = 10.62DD251 pKa = 3.61SNGIADD257 pKa = 3.72TAYY260 pKa = 9.58ISVNGVRR267 pKa = 11.84TTGIEE272 pKa = 3.89NTVYY276 pKa = 10.88TNDD279 pKa = 3.93AQPLPYY285 pKa = 10.12FIRR288 pKa = 11.84ALDD291 pKa = 3.64SKK293 pKa = 7.9YY294 pKa = 8.08TWGAIGQGQTLNYY307 pKa = 10.32SFALNADD314 pKa = 3.41GGEE317 pKa = 4.31VGFEE321 pKa = 4.68LYY323 pKa = 10.14TEE325 pKa = 4.17EE326 pKa = 3.86QKK328 pKa = 9.8TAIRR332 pKa = 11.84EE333 pKa = 4.14ALQKK337 pKa = 10.37YY338 pKa = 8.63AQLVNLTFVEE348 pKa = 4.93VVDD351 pKa = 4.0STSVQLRR358 pKa = 11.84FFRR361 pKa = 11.84DD362 pKa = 3.56DD363 pKa = 3.85LSTAGYY369 pKa = 9.49AAYY372 pKa = 9.91AGYY375 pKa = 11.18ANYY378 pKa = 9.94PFDD381 pKa = 3.8SSKK384 pKa = 11.36GGDD387 pKa = 2.93IHH389 pKa = 7.72IKK391 pKa = 8.82NTISNLNQLDD401 pKa = 3.64GSFDD405 pKa = 3.74VLIHH409 pKa = 6.6EE410 pKa = 5.04IGHH413 pKa = 6.26ALGLKK418 pKa = 10.24HH419 pKa = 6.57PFEE422 pKa = 5.07DD423 pKa = 4.07TDD425 pKa = 3.91TLPTSEE431 pKa = 6.04DD432 pKa = 3.48STTYY436 pKa = 11.12SAMSYY441 pKa = 10.59NSLWQANKK449 pKa = 8.67NTVRR453 pKa = 11.84MYY455 pKa = 11.38DD456 pKa = 3.65LATLHH461 pKa = 6.09YY462 pKa = 9.38FYY464 pKa = 11.04GVNASQRR471 pKa = 11.84SEE473 pKa = 3.8NNTYY477 pKa = 11.08AFSDD481 pKa = 3.72ANFTDD486 pKa = 3.95HH487 pKa = 6.65YY488 pKa = 9.3VWDD491 pKa = 4.19GAGVDD496 pKa = 3.77VFDD499 pKa = 6.52ANSQTQNVSVNLNPGSWNFIGSKK522 pKa = 10.07AVSILSAGQSFIGFGTLIEE541 pKa = 4.25NAVSGSGNDD550 pKa = 3.67TVIGNALDD558 pKa = 3.97NQIHH562 pKa = 5.93TNAGNDD568 pKa = 3.49WLKK571 pKa = 11.29GGAGNDD577 pKa = 3.18SLYY580 pKa = 11.04AGEE583 pKa = 5.88GNDD586 pKa = 3.74TLNGQVGADD595 pKa = 3.42VMVGGNGDD603 pKa = 3.44DD604 pKa = 3.94YY605 pKa = 11.63YY606 pKa = 11.84YY607 pKa = 10.89FDD609 pKa = 3.9NVGDD613 pKa = 3.91AVIEE617 pKa = 4.23VVDD620 pKa = 4.23AGLDD624 pKa = 3.75CVEE627 pKa = 4.93ASANCTLNDD636 pKa = 3.79NIEE639 pKa = 4.02NLVLSGSGALSGMGNALNNQMKK661 pKa = 10.52GSAEE665 pKa = 4.0NNTLKK670 pKa = 10.82GMAGNDD676 pKa = 3.52SLLGFAGNDD685 pKa = 3.45ALDD688 pKa = 4.11GGTGNDD694 pKa = 3.63TLRR697 pKa = 11.84GGLGNDD703 pKa = 3.52VLNGGDD709 pKa = 3.82GIDD712 pKa = 3.28WADD715 pKa = 3.82YY716 pKa = 11.1VDD718 pKa = 3.89MTASVNVNLNITTVQVTGAGGSDD741 pKa = 3.08TLTNIEE747 pKa = 4.08NVRR750 pKa = 11.84AGSANDD756 pKa = 3.54NLKK759 pKa = 11.11GNASDD764 pKa = 4.61NVLDD768 pKa = 4.29GQAGNDD774 pKa = 3.48ALFGNAGNDD783 pKa = 3.55NLIGGAGNDD792 pKa = 3.6ILNGGLGNDD801 pKa = 3.86NLSGGDD807 pKa = 3.57GNDD810 pKa = 2.99WADD813 pKa = 3.78YY814 pKa = 10.22FDD816 pKa = 3.51STAAVSVDD824 pKa = 3.57LRR826 pKa = 11.84LATQQNTVGAGLDD839 pKa = 2.89IFYY842 pKa = 10.52NIEE845 pKa = 3.91NLRR848 pKa = 11.84GGLGNDD854 pKa = 3.6TLNGSVYY861 pKa = 11.12ANALVGQAGNDD872 pKa = 3.35FMVGNGGNDD881 pKa = 3.12TLYY884 pKa = 11.05AGLGNDD890 pKa = 3.71TLFGGTGNDD899 pKa = 2.84TYY901 pKa = 11.38QFNRR905 pKa = 11.84GDD907 pKa = 3.93GQDD910 pKa = 3.41TLTDD914 pKa = 3.4ADD916 pKa = 3.79ATVGNSDD923 pKa = 3.47TLAFQTGVTNSQLWLTHH940 pKa = 5.94TGNDD944 pKa = 3.35LVVSVIGTTDD954 pKa = 2.87KK955 pKa = 10.7VTIKK959 pKa = 10.26NWYY962 pKa = 8.22VSSANHH968 pKa = 5.79VEE970 pKa = 4.34SITAGGKK977 pKa = 7.53TLADD981 pKa = 3.73GNVNALVAAMSAFSPPAAGQTTLPSSYY1008 pKa = 8.48QTALNPVIAANWVV1021 pKa = 3.13
Molecular weight: 106.7 kDa
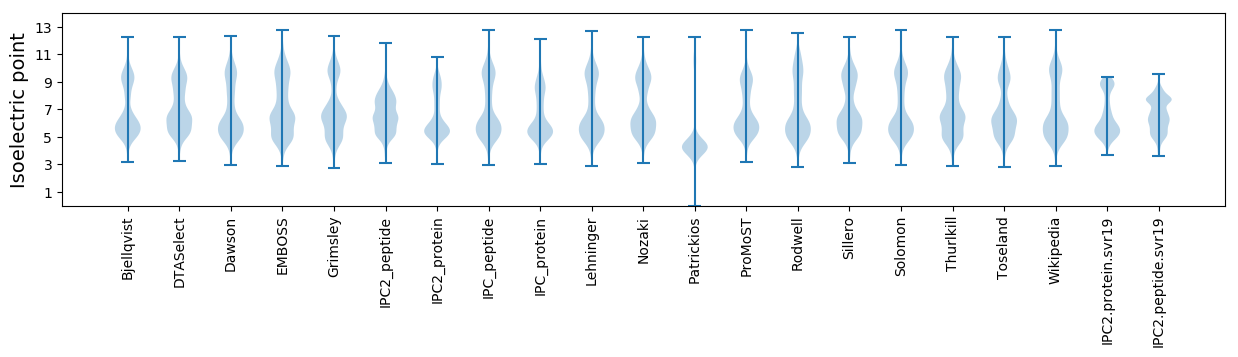
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345DDD9|A0A345DDD9_9BURK 2-aminoadipate transaminase OS=Ephemeroptericola cinctiostellae OX=2268024 GN=lysN_2 PE=4 SV=1
MM1 pKa = 7.6AAPRR5 pKa = 11.84SKK7 pKa = 9.47TGRR10 pKa = 11.84PEE12 pKa = 3.46RR13 pKa = 11.84RR14 pKa = 11.84PAQNPLFKK22 pKa = 10.35RR23 pKa = 11.84KK24 pKa = 8.99RR25 pKa = 11.84VCRR28 pKa = 11.84FTMAKK33 pKa = 9.68VDD35 pKa = 3.74QIDD38 pKa = 3.91YY39 pKa = 11.04KK40 pKa = 11.33DD41 pKa = 3.47VDD43 pKa = 3.83TLKK46 pKa = 11.18DD47 pKa = 4.79FIAEE51 pKa = 3.96NGKK54 pKa = 10.03IIPARR59 pKa = 11.84LTGTRR64 pKa = 11.84AIYY67 pKa = 9.74QRR69 pKa = 11.84QLNTCIKK76 pKa = 9.93RR77 pKa = 11.84ARR79 pKa = 11.84FLALVPYY86 pKa = 9.78CDD88 pKa = 3.37NHH90 pKa = 6.44
MM1 pKa = 7.6AAPRR5 pKa = 11.84SKK7 pKa = 9.47TGRR10 pKa = 11.84PEE12 pKa = 3.46RR13 pKa = 11.84RR14 pKa = 11.84PAQNPLFKK22 pKa = 10.35RR23 pKa = 11.84KK24 pKa = 8.99RR25 pKa = 11.84VCRR28 pKa = 11.84FTMAKK33 pKa = 9.68VDD35 pKa = 3.74QIDD38 pKa = 3.91YY39 pKa = 11.04KK40 pKa = 11.33DD41 pKa = 3.47VDD43 pKa = 3.83TLKK46 pKa = 11.18DD47 pKa = 4.79FIAEE51 pKa = 3.96NGKK54 pKa = 10.03IIPARR59 pKa = 11.84LTGTRR64 pKa = 11.84AIYY67 pKa = 9.74QRR69 pKa = 11.84QLNTCIKK76 pKa = 9.93RR77 pKa = 11.84ARR79 pKa = 11.84FLALVPYY86 pKa = 9.78CDD88 pKa = 3.37NHH90 pKa = 6.44
Molecular weight: 10.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
776007 |
30 |
4882 |
313.4 |
34.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.571 ± 0.069 | 0.973 ± 0.018 |
5.556 ± 0.052 | 5.439 ± 0.106 |
3.901 ± 0.041 | 7.117 ± 0.082 |
2.665 ± 0.04 | 5.573 ± 0.043 |
4.6 ± 0.045 | 10.085 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.864 ± 0.031 | 4.042 ± 0.04 |
4.228 ± 0.039 | 4.106 ± 0.049 |
4.838 ± 0.056 | 5.842 ± 0.047 |
5.729 ± 0.047 | 7.875 ± 0.127 |
1.347 ± 0.026 | 2.651 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
