
Penicillium brevicompactum tetramycovirus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus; Penicillum brevicompactum polymycovirus 1
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
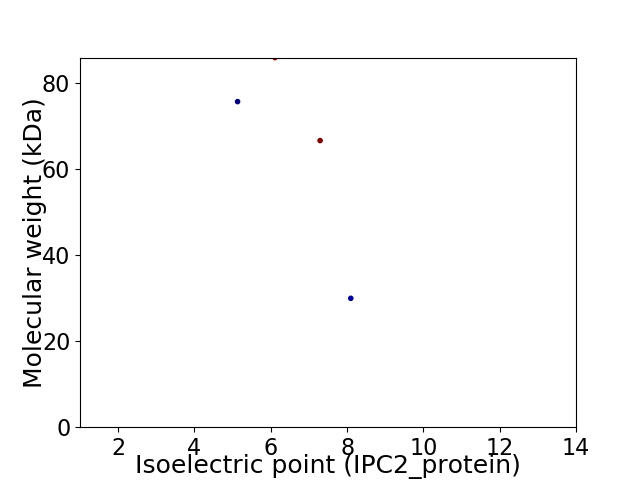
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A3G3C4P0|A0A3G3C4P0_9VIRU RNA-dependent RNA polymerase OS=Penicillium brevicompactum tetramycovirus 1 OX=2485923 PE=4 SV=1
MM1 pKa = 8.02ADD3 pKa = 3.73LTRR6 pKa = 11.84LASIALNVDD15 pKa = 3.43SPAATLAAVVYY26 pKa = 7.15YY27 pKa = 9.31TGKK30 pKa = 8.02VTPGAEE36 pKa = 4.11YY37 pKa = 10.93VEE39 pKa = 4.41PAVLRR44 pKa = 11.84DD45 pKa = 3.55VLGWIAASPMLSIGNVVNEE64 pKa = 3.93EE65 pKa = 3.45AVAYY69 pKa = 9.77VRR71 pKa = 11.84LYY73 pKa = 10.94NGSDD77 pKa = 3.08KK78 pKa = 11.37LNNSVLDD85 pKa = 3.91DD86 pKa = 3.64ASYY89 pKa = 10.57YY90 pKa = 10.37LRR92 pKa = 11.84HH93 pKa = 6.42FGPLGGSYY101 pKa = 9.91TKK103 pKa = 10.34DD104 pKa = 2.84ARR106 pKa = 11.84TVRR109 pKa = 11.84VAEE112 pKa = 4.71IINHH116 pKa = 5.91ASIHH120 pKa = 5.39QGEE123 pKa = 4.72ATTGPVEE130 pKa = 4.13TALLEE135 pKa = 4.44SVSVAGYY142 pKa = 9.88EE143 pKa = 4.25GLAGALGAVSRR154 pKa = 11.84LHH156 pKa = 5.78LQQVIDD162 pKa = 3.96KK163 pKa = 9.13PDD165 pKa = 3.39VVVTQLDD172 pKa = 3.63RR173 pKa = 11.84AVFTASTVARR183 pKa = 11.84EE184 pKa = 4.04ATGEE188 pKa = 4.03HH189 pKa = 6.7RR190 pKa = 11.84FLATPPCVSRR200 pKa = 11.84QSAIACLAVMLSVTHH215 pKa = 6.56RR216 pKa = 11.84RR217 pKa = 11.84LVTDD221 pKa = 4.97RR222 pKa = 11.84IRR224 pKa = 11.84EE225 pKa = 3.97KK226 pKa = 10.32ATTVTSMAARR236 pKa = 11.84IDD238 pKa = 3.78LEE240 pKa = 4.02AVRR243 pKa = 11.84DD244 pKa = 3.65IVYY247 pKa = 8.56PARR250 pKa = 11.84EE251 pKa = 4.65LITNYY256 pKa = 9.75IFDD259 pKa = 4.29PAEE262 pKa = 3.86LVLRR266 pKa = 11.84DD267 pKa = 3.19QNGRR271 pKa = 11.84VRR273 pKa = 11.84EE274 pKa = 4.12RR275 pKa = 11.84ATSRR279 pKa = 11.84SPAAIVAFAFLTSRR293 pKa = 11.84GSSDD297 pKa = 3.82VLMDD301 pKa = 4.13LANRR305 pKa = 11.84RR306 pKa = 11.84VDD308 pKa = 3.55AVVDD312 pKa = 3.68KK313 pKa = 11.13DD314 pKa = 3.71SLVPHH319 pKa = 5.21VQEE322 pKa = 4.85YY323 pKa = 10.15YY324 pKa = 10.75DD325 pKa = 3.63NSRR328 pKa = 11.84TIYY331 pKa = 10.66SSLLLFLRR339 pKa = 11.84ALEE342 pKa = 4.05GTRR345 pKa = 11.84WEE347 pKa = 4.12RR348 pKa = 11.84ANNGRR353 pKa = 11.84GEE355 pKa = 4.31DD356 pKa = 3.97VNKK359 pKa = 10.34GPCLGYY365 pKa = 9.18MAAGRR370 pKa = 11.84TRR372 pKa = 11.84RR373 pKa = 11.84DD374 pKa = 3.19RR375 pKa = 11.84NTAAAVNRR383 pKa = 11.84LSEE386 pKa = 4.21MVVEE390 pKa = 4.16NRR392 pKa = 11.84EE393 pKa = 4.16LVPEE397 pKa = 4.81AGRR400 pKa = 11.84ILYY403 pKa = 9.92ILDD406 pKa = 3.18WNGVYY411 pKa = 10.26DD412 pKa = 4.09RR413 pKa = 11.84SVVLGAASLMKK424 pKa = 9.85IDD426 pKa = 3.79VALDD430 pKa = 3.35MGASGHH436 pKa = 6.67DD437 pKa = 3.5VGGVANPDD445 pKa = 3.41KK446 pKa = 11.0ASQTRR451 pKa = 11.84HH452 pKa = 4.22YY453 pKa = 9.63TVLLPALPGRR463 pKa = 11.84SMPTCSTVEE472 pKa = 3.97YY473 pKa = 10.7QPGDD477 pKa = 3.33GTASRR482 pKa = 11.84IRR484 pKa = 11.84ILVDD488 pKa = 4.51AIGGFNDD495 pKa = 3.87AYY497 pKa = 10.93SYY499 pKa = 10.88IYY501 pKa = 10.71GGTPADD507 pKa = 3.72KK508 pKa = 10.15TITPAVSVDD517 pKa = 3.22EE518 pKa = 4.38AQARR522 pKa = 11.84VFAVSGSSVGIPNALSTVDD541 pKa = 3.49ILLPPSCYY549 pKa = 9.53HH550 pKa = 6.43WPATPVDD557 pKa = 3.8TMVDD561 pKa = 3.25TFGDD565 pKa = 3.47VDD567 pKa = 4.41EE568 pKa = 5.81SCMSCQDD575 pKa = 2.97SVRR578 pKa = 11.84ALRR581 pKa = 11.84RR582 pKa = 11.84TAEE585 pKa = 4.14LLSVVGARR593 pKa = 11.84LVKK596 pKa = 8.74TRR598 pKa = 11.84SAFGHH603 pKa = 5.92NAQFDD608 pKa = 4.28IEE610 pKa = 4.45VSPAIRR616 pKa = 11.84DD617 pKa = 3.75GASDD621 pKa = 3.42TGVTLDD627 pKa = 3.34ATAMLNHH634 pKa = 6.17MRR636 pKa = 11.84NKK638 pKa = 10.43DD639 pKa = 3.14WGGDD643 pKa = 3.4TFRR646 pKa = 11.84PGGVVASTNQYY657 pKa = 7.6TAEE660 pKa = 3.9VDD662 pKa = 4.18EE663 pKa = 4.79EE664 pKa = 4.33CHH666 pKa = 7.27AILTHH671 pKa = 7.06IRR673 pKa = 11.84MLTYY677 pKa = 10.63EE678 pKa = 4.44DD679 pKa = 3.66LAGIGEE685 pKa = 4.47AAGPLPSSDD694 pKa = 3.61FDD696 pKa = 3.97DD697 pKa = 3.92VARR700 pKa = 11.84SLGG703 pKa = 3.51
MM1 pKa = 8.02ADD3 pKa = 3.73LTRR6 pKa = 11.84LASIALNVDD15 pKa = 3.43SPAATLAAVVYY26 pKa = 7.15YY27 pKa = 9.31TGKK30 pKa = 8.02VTPGAEE36 pKa = 4.11YY37 pKa = 10.93VEE39 pKa = 4.41PAVLRR44 pKa = 11.84DD45 pKa = 3.55VLGWIAASPMLSIGNVVNEE64 pKa = 3.93EE65 pKa = 3.45AVAYY69 pKa = 9.77VRR71 pKa = 11.84LYY73 pKa = 10.94NGSDD77 pKa = 3.08KK78 pKa = 11.37LNNSVLDD85 pKa = 3.91DD86 pKa = 3.64ASYY89 pKa = 10.57YY90 pKa = 10.37LRR92 pKa = 11.84HH93 pKa = 6.42FGPLGGSYY101 pKa = 9.91TKK103 pKa = 10.34DD104 pKa = 2.84ARR106 pKa = 11.84TVRR109 pKa = 11.84VAEE112 pKa = 4.71IINHH116 pKa = 5.91ASIHH120 pKa = 5.39QGEE123 pKa = 4.72ATTGPVEE130 pKa = 4.13TALLEE135 pKa = 4.44SVSVAGYY142 pKa = 9.88EE143 pKa = 4.25GLAGALGAVSRR154 pKa = 11.84LHH156 pKa = 5.78LQQVIDD162 pKa = 3.96KK163 pKa = 9.13PDD165 pKa = 3.39VVVTQLDD172 pKa = 3.63RR173 pKa = 11.84AVFTASTVARR183 pKa = 11.84EE184 pKa = 4.04ATGEE188 pKa = 4.03HH189 pKa = 6.7RR190 pKa = 11.84FLATPPCVSRR200 pKa = 11.84QSAIACLAVMLSVTHH215 pKa = 6.56RR216 pKa = 11.84RR217 pKa = 11.84LVTDD221 pKa = 4.97RR222 pKa = 11.84IRR224 pKa = 11.84EE225 pKa = 3.97KK226 pKa = 10.32ATTVTSMAARR236 pKa = 11.84IDD238 pKa = 3.78LEE240 pKa = 4.02AVRR243 pKa = 11.84DD244 pKa = 3.65IVYY247 pKa = 8.56PARR250 pKa = 11.84EE251 pKa = 4.65LITNYY256 pKa = 9.75IFDD259 pKa = 4.29PAEE262 pKa = 3.86LVLRR266 pKa = 11.84DD267 pKa = 3.19QNGRR271 pKa = 11.84VRR273 pKa = 11.84EE274 pKa = 4.12RR275 pKa = 11.84ATSRR279 pKa = 11.84SPAAIVAFAFLTSRR293 pKa = 11.84GSSDD297 pKa = 3.82VLMDD301 pKa = 4.13LANRR305 pKa = 11.84RR306 pKa = 11.84VDD308 pKa = 3.55AVVDD312 pKa = 3.68KK313 pKa = 11.13DD314 pKa = 3.71SLVPHH319 pKa = 5.21VQEE322 pKa = 4.85YY323 pKa = 10.15YY324 pKa = 10.75DD325 pKa = 3.63NSRR328 pKa = 11.84TIYY331 pKa = 10.66SSLLLFLRR339 pKa = 11.84ALEE342 pKa = 4.05GTRR345 pKa = 11.84WEE347 pKa = 4.12RR348 pKa = 11.84ANNGRR353 pKa = 11.84GEE355 pKa = 4.31DD356 pKa = 3.97VNKK359 pKa = 10.34GPCLGYY365 pKa = 9.18MAAGRR370 pKa = 11.84TRR372 pKa = 11.84RR373 pKa = 11.84DD374 pKa = 3.19RR375 pKa = 11.84NTAAAVNRR383 pKa = 11.84LSEE386 pKa = 4.21MVVEE390 pKa = 4.16NRR392 pKa = 11.84EE393 pKa = 4.16LVPEE397 pKa = 4.81AGRR400 pKa = 11.84ILYY403 pKa = 9.92ILDD406 pKa = 3.18WNGVYY411 pKa = 10.26DD412 pKa = 4.09RR413 pKa = 11.84SVVLGAASLMKK424 pKa = 9.85IDD426 pKa = 3.79VALDD430 pKa = 3.35MGASGHH436 pKa = 6.67DD437 pKa = 3.5VGGVANPDD445 pKa = 3.41KK446 pKa = 11.0ASQTRR451 pKa = 11.84HH452 pKa = 4.22YY453 pKa = 9.63TVLLPALPGRR463 pKa = 11.84SMPTCSTVEE472 pKa = 3.97YY473 pKa = 10.7QPGDD477 pKa = 3.33GTASRR482 pKa = 11.84IRR484 pKa = 11.84ILVDD488 pKa = 4.51AIGGFNDD495 pKa = 3.87AYY497 pKa = 10.93SYY499 pKa = 10.88IYY501 pKa = 10.71GGTPADD507 pKa = 3.72KK508 pKa = 10.15TITPAVSVDD517 pKa = 3.22EE518 pKa = 4.38AQARR522 pKa = 11.84VFAVSGSSVGIPNALSTVDD541 pKa = 3.49ILLPPSCYY549 pKa = 9.53HH550 pKa = 6.43WPATPVDD557 pKa = 3.8TMVDD561 pKa = 3.25TFGDD565 pKa = 3.47VDD567 pKa = 4.41EE568 pKa = 5.81SCMSCQDD575 pKa = 2.97SVRR578 pKa = 11.84ALRR581 pKa = 11.84RR582 pKa = 11.84TAEE585 pKa = 4.14LLSVVGARR593 pKa = 11.84LVKK596 pKa = 8.74TRR598 pKa = 11.84SAFGHH603 pKa = 5.92NAQFDD608 pKa = 4.28IEE610 pKa = 4.45VSPAIRR616 pKa = 11.84DD617 pKa = 3.75GASDD621 pKa = 3.42TGVTLDD627 pKa = 3.34ATAMLNHH634 pKa = 6.17MRR636 pKa = 11.84NKK638 pKa = 10.43DD639 pKa = 3.14WGGDD643 pKa = 3.4TFRR646 pKa = 11.84PGGVVASTNQYY657 pKa = 7.6TAEE660 pKa = 3.9VDD662 pKa = 4.18EE663 pKa = 4.79EE664 pKa = 4.33CHH666 pKa = 7.27AILTHH671 pKa = 7.06IRR673 pKa = 11.84MLTYY677 pKa = 10.63EE678 pKa = 4.44DD679 pKa = 3.66LAGIGEE685 pKa = 4.47AAGPLPSSDD694 pKa = 3.61FDD696 pKa = 3.97DD697 pKa = 3.92VARR700 pKa = 11.84SLGG703 pKa = 3.51
Molecular weight: 75.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G3C4W9|A0A3G3C4W9_9VIRU Methyltransferase OS=Penicillium brevicompactum tetramycovirus 1 OX=2485923 PE=4 SV=1
MM1 pKa = 7.43SSHH4 pKa = 6.18VGKK7 pKa = 10.23IASFLSTADD16 pKa = 3.49LSRR19 pKa = 11.84VRR21 pKa = 11.84TLNNEE26 pKa = 3.77DD27 pKa = 3.1VQLVVRR33 pKa = 11.84CLAAGVPPKK42 pKa = 10.83AFAEE46 pKa = 4.41TLGSYY51 pKa = 10.37DD52 pKa = 3.45PNKK55 pKa = 10.43EE56 pKa = 3.8YY57 pKa = 11.02DD58 pKa = 3.9VLPSAGPVPTRR69 pKa = 11.84KK70 pKa = 9.31ILAYY74 pKa = 10.36SFLRR78 pKa = 11.84DD79 pKa = 3.06RR80 pKa = 11.84RR81 pKa = 11.84QYY83 pKa = 11.6ADD85 pKa = 3.55TYY87 pKa = 10.06AQSDD91 pKa = 3.5EE92 pKa = 4.0WAGRR96 pKa = 11.84LRR98 pKa = 11.84GLLDD102 pKa = 3.81TDD104 pKa = 3.55PKK106 pKa = 10.71KK107 pKa = 10.85AVDD110 pKa = 4.35EE111 pKa = 4.39ISTVVNRR118 pKa = 11.84RR119 pKa = 11.84LRR121 pKa = 11.84VKK123 pKa = 10.57GSPAVADD130 pKa = 3.67VEE132 pKa = 4.65VGQVRR137 pKa = 11.84GSTGSEE143 pKa = 3.76ATGGASDD150 pKa = 4.46PNYY153 pKa = 11.1APLQQAMLKK162 pKa = 10.36YY163 pKa = 9.81SAQAGAFKK171 pKa = 10.39FDD173 pKa = 4.09AADD176 pKa = 3.69CGTPRR181 pKa = 11.84DD182 pKa = 3.55RR183 pKa = 11.84RR184 pKa = 11.84FVVTLGARR192 pKa = 11.84LIVGAQKK199 pKa = 10.81KK200 pKa = 7.98STAIMAARR208 pKa = 11.84LCRR211 pKa = 11.84VLGRR215 pKa = 11.84HH216 pKa = 6.12DD217 pKa = 4.2RR218 pKa = 11.84VVSPYY223 pKa = 10.0VGHH226 pKa = 6.61VDD228 pKa = 3.78EE229 pKa = 5.74EE230 pKa = 4.66EE231 pKa = 3.89DD232 pKa = 4.17HH233 pKa = 6.96EE234 pKa = 4.38WDD236 pKa = 3.55EE237 pKa = 4.98EE238 pKa = 4.34IPADD242 pKa = 3.85VADD245 pKa = 4.77GKK247 pKa = 9.46PVRR250 pKa = 11.84TADD253 pKa = 3.82RR254 pKa = 11.84AKK256 pKa = 10.89SPTTTRR262 pKa = 11.84SATPTSPKK270 pKa = 10.43GGAAGAAGGKK280 pKa = 9.37
MM1 pKa = 7.43SSHH4 pKa = 6.18VGKK7 pKa = 10.23IASFLSTADD16 pKa = 3.49LSRR19 pKa = 11.84VRR21 pKa = 11.84TLNNEE26 pKa = 3.77DD27 pKa = 3.1VQLVVRR33 pKa = 11.84CLAAGVPPKK42 pKa = 10.83AFAEE46 pKa = 4.41TLGSYY51 pKa = 10.37DD52 pKa = 3.45PNKK55 pKa = 10.43EE56 pKa = 3.8YY57 pKa = 11.02DD58 pKa = 3.9VLPSAGPVPTRR69 pKa = 11.84KK70 pKa = 9.31ILAYY74 pKa = 10.36SFLRR78 pKa = 11.84DD79 pKa = 3.06RR80 pKa = 11.84RR81 pKa = 11.84QYY83 pKa = 11.6ADD85 pKa = 3.55TYY87 pKa = 10.06AQSDD91 pKa = 3.5EE92 pKa = 4.0WAGRR96 pKa = 11.84LRR98 pKa = 11.84GLLDD102 pKa = 3.81TDD104 pKa = 3.55PKK106 pKa = 10.71KK107 pKa = 10.85AVDD110 pKa = 4.35EE111 pKa = 4.39ISTVVNRR118 pKa = 11.84RR119 pKa = 11.84LRR121 pKa = 11.84VKK123 pKa = 10.57GSPAVADD130 pKa = 3.67VEE132 pKa = 4.65VGQVRR137 pKa = 11.84GSTGSEE143 pKa = 3.76ATGGASDD150 pKa = 4.46PNYY153 pKa = 11.1APLQQAMLKK162 pKa = 10.36YY163 pKa = 9.81SAQAGAFKK171 pKa = 10.39FDD173 pKa = 4.09AADD176 pKa = 3.69CGTPRR181 pKa = 11.84DD182 pKa = 3.55RR183 pKa = 11.84RR184 pKa = 11.84FVVTLGARR192 pKa = 11.84LIVGAQKK199 pKa = 10.81KK200 pKa = 7.98STAIMAARR208 pKa = 11.84LCRR211 pKa = 11.84VLGRR215 pKa = 11.84HH216 pKa = 6.12DD217 pKa = 4.2RR218 pKa = 11.84VVSPYY223 pKa = 10.0VGHH226 pKa = 6.61VDD228 pKa = 3.78EE229 pKa = 5.74EE230 pKa = 4.66EE231 pKa = 3.89DD232 pKa = 4.17HH233 pKa = 6.96EE234 pKa = 4.38WDD236 pKa = 3.55EE237 pKa = 4.98EE238 pKa = 4.34IPADD242 pKa = 3.85VADD245 pKa = 4.77GKK247 pKa = 9.46PVRR250 pKa = 11.84TADD253 pKa = 3.82RR254 pKa = 11.84AKK256 pKa = 10.89SPTTTRR262 pKa = 11.84SATPTSPKK270 pKa = 10.43GGAAGAAGGKK280 pKa = 9.37
Molecular weight: 29.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2370 |
280 |
775 |
592.5 |
64.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.549 ± 0.827 | 1.308 ± 0.098 |
7.046 ± 0.173 | 4.937 ± 0.338 |
3.333 ± 0.557 | 7.553 ± 0.262 |
2.363 ± 0.224 | 3.882 ± 0.278 |
3.502 ± 0.597 | 8.354 ± 0.301 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.321 ± 0.39 | 2.616 ± 0.308 |
5.485 ± 0.547 | 2.616 ± 0.38 |
7.764 ± 0.234 | 7.384 ± 0.306 |
5.738 ± 0.436 | 9.705 ± 0.659 |
0.844 ± 0.056 | 2.7 ± 0.306 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
