
Halioglobus sp. HI00S01
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Halioglobus; unclassified Halioglobus
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
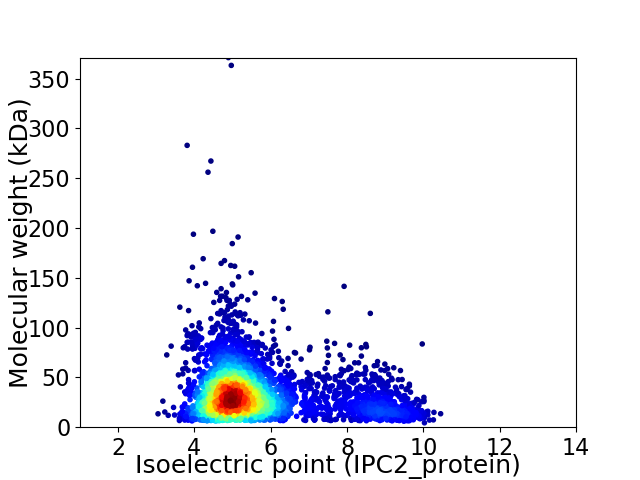
Virtual 2D-PAGE plot for 4067 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A162H7Y7|A0A162H7Y7_9GAMM Uncharacterized protein OS=Halioglobus sp. HI00S01 OX=1822214 GN=A3709_00840 PE=4 SV=1
MM1 pKa = 6.86ITRR4 pKa = 11.84FASLSSRR11 pKa = 11.84LAATLIALCAITACGGGGGGGGGGGFLGDD40 pKa = 4.02SGGSSDD46 pKa = 3.81TYY48 pKa = 10.62FIQVALKK55 pKa = 10.69DD56 pKa = 3.53SDD58 pKa = 4.03GNEE61 pKa = 4.19TSTVTPNSPGTLQVLVTQNNPNGNPIADD89 pKa = 4.14VIVTASTNSGLLFPSSGSKK108 pKa = 8.48LTNAEE113 pKa = 3.75GAVTFRR119 pKa = 11.84IEE121 pKa = 4.04AGNEE125 pKa = 3.62SRR127 pKa = 11.84GAGTIEE133 pKa = 4.19VSVEE137 pKa = 3.9DD138 pKa = 3.73PSGNVVTEE146 pKa = 4.36SINFQLGVNGLRR158 pKa = 11.84LGFILDD164 pKa = 3.58GVYY167 pKa = 10.53YY168 pKa = 10.47DD169 pKa = 4.49AEE171 pKa = 4.37IGIAPNEE178 pKa = 4.39PIAAQGSAILALAVVDD194 pKa = 4.41EE195 pKa = 4.64NDD197 pKa = 3.44VPVAGAEE204 pKa = 4.33SVRR207 pKa = 11.84ITSACINSGEE217 pKa = 4.38STLSPEE223 pKa = 4.38NPIAVPNGQITVDD236 pKa = 3.88YY237 pKa = 10.81QVGSCSGIDD246 pKa = 3.3QLTAEE251 pKa = 4.92IIGTEE256 pKa = 3.84AQAFGTIEE264 pKa = 3.96IAEE267 pKa = 4.28PAADD271 pKa = 3.44SLTFISADD279 pKa = 3.13PTLIVLRR286 pKa = 11.84GTGGGPDD293 pKa = 2.9RR294 pKa = 11.84KK295 pKa = 9.66EE296 pKa = 3.56QSEE299 pKa = 4.79VKK301 pKa = 10.51FEE303 pKa = 4.31AVDD306 pKa = 3.66SQGNPQEE313 pKa = 4.07GVMVDD318 pKa = 4.46FSLTTDD324 pKa = 3.01VGGITFNPASATTDD338 pKa = 2.99AEE340 pKa = 4.97GIVSTFVNSGDD351 pKa = 3.32VATVVRR357 pKa = 11.84VVATMDD363 pKa = 3.14AMNGEE368 pKa = 4.57GTVSAVSDD376 pKa = 3.72VLTISTGLPDD386 pKa = 3.58QNSISLSVEE395 pKa = 3.81GTSVMIEE402 pKa = 3.63DD403 pKa = 4.09AMRR406 pKa = 11.84VDD408 pKa = 5.08GITRR412 pKa = 11.84AITVRR417 pKa = 11.84MADD420 pKa = 3.31KK421 pKa = 10.68FNNPVPDD428 pKa = 3.34GTAAVFTTEE437 pKa = 3.94YY438 pKa = 11.14GSIEE442 pKa = 4.41SACQTTNGACSVTWTSQAPRR462 pKa = 11.84FPTLPEE468 pKa = 3.78NQEE471 pKa = 4.21LVRR474 pKa = 11.84TIYY477 pKa = 10.81DD478 pKa = 3.18ADD480 pKa = 4.04YY481 pKa = 11.25ACPSHH486 pKa = 6.25NANGGPCPDD495 pKa = 3.94DD496 pKa = 4.01LGFIRR501 pKa = 11.84GARR504 pKa = 11.84STILVTAIGEE514 pKa = 4.15EE515 pKa = 4.25SFIDD519 pKa = 3.59ANANGIYY526 pKa = 10.27DD527 pKa = 3.61EE528 pKa = 4.78GEE530 pKa = 3.73RR531 pKa = 11.84FANLTEE537 pKa = 4.7AFLDD541 pKa = 3.99NNWNGFYY548 pKa = 10.9DD549 pKa = 3.84PARR552 pKa = 11.84TYY554 pKa = 10.81CVNNPTLRR562 pKa = 11.84EE563 pKa = 3.9CRR565 pKa = 11.84AGSEE569 pKa = 4.61EE570 pKa = 4.94IFTDD574 pKa = 4.36FNNNGVFDD582 pKa = 5.21ANGDD586 pKa = 3.89DD587 pKa = 4.23PANGYY592 pKa = 9.15PDD594 pKa = 3.69EE595 pKa = 4.88GVTAVYY601 pKa = 10.6NGLLCPPEE609 pKa = 5.79GDD611 pKa = 3.2GVYY614 pKa = 10.44CSRR617 pKa = 11.84EE618 pKa = 4.03LVNVRR623 pKa = 11.84DD624 pKa = 4.26DD625 pKa = 3.83LQIILSDD632 pKa = 3.64NPAWDD637 pKa = 3.48VLVVDD642 pKa = 4.06NRR644 pKa = 11.84GNAVTDD650 pKa = 3.38TAFNGGEE657 pKa = 4.05YY658 pKa = 9.73QVYY661 pKa = 10.23VSDD664 pKa = 4.17TFNNAPIAGTTISVTSSVCEE684 pKa = 3.98VNLSGNTEE692 pKa = 3.99VPNAILPGAFGVSLTQGGSVTYY714 pKa = 10.26ASCRR718 pKa = 11.84DD719 pKa = 3.39EE720 pKa = 4.42EE721 pKa = 4.55RR722 pKa = 11.84PSNTSEE728 pKa = 3.7FQINVGSEE736 pKa = 3.96YY737 pKa = 11.03NEE739 pKa = 4.12TFSCVIQVIDD749 pKa = 4.75DD750 pKa = 4.32ASDD753 pKa = 3.61CPEE756 pKa = 4.35PEE758 pKa = 4.57GGGGGDD764 pKa = 4.45DD765 pKa = 3.76GLSNGGG771 pKa = 3.49
MM1 pKa = 6.86ITRR4 pKa = 11.84FASLSSRR11 pKa = 11.84LAATLIALCAITACGGGGGGGGGGGFLGDD40 pKa = 4.02SGGSSDD46 pKa = 3.81TYY48 pKa = 10.62FIQVALKK55 pKa = 10.69DD56 pKa = 3.53SDD58 pKa = 4.03GNEE61 pKa = 4.19TSTVTPNSPGTLQVLVTQNNPNGNPIADD89 pKa = 4.14VIVTASTNSGLLFPSSGSKK108 pKa = 8.48LTNAEE113 pKa = 3.75GAVTFRR119 pKa = 11.84IEE121 pKa = 4.04AGNEE125 pKa = 3.62SRR127 pKa = 11.84GAGTIEE133 pKa = 4.19VSVEE137 pKa = 3.9DD138 pKa = 3.73PSGNVVTEE146 pKa = 4.36SINFQLGVNGLRR158 pKa = 11.84LGFILDD164 pKa = 3.58GVYY167 pKa = 10.53YY168 pKa = 10.47DD169 pKa = 4.49AEE171 pKa = 4.37IGIAPNEE178 pKa = 4.39PIAAQGSAILALAVVDD194 pKa = 4.41EE195 pKa = 4.64NDD197 pKa = 3.44VPVAGAEE204 pKa = 4.33SVRR207 pKa = 11.84ITSACINSGEE217 pKa = 4.38STLSPEE223 pKa = 4.38NPIAVPNGQITVDD236 pKa = 3.88YY237 pKa = 10.81QVGSCSGIDD246 pKa = 3.3QLTAEE251 pKa = 4.92IIGTEE256 pKa = 3.84AQAFGTIEE264 pKa = 3.96IAEE267 pKa = 4.28PAADD271 pKa = 3.44SLTFISADD279 pKa = 3.13PTLIVLRR286 pKa = 11.84GTGGGPDD293 pKa = 2.9RR294 pKa = 11.84KK295 pKa = 9.66EE296 pKa = 3.56QSEE299 pKa = 4.79VKK301 pKa = 10.51FEE303 pKa = 4.31AVDD306 pKa = 3.66SQGNPQEE313 pKa = 4.07GVMVDD318 pKa = 4.46FSLTTDD324 pKa = 3.01VGGITFNPASATTDD338 pKa = 2.99AEE340 pKa = 4.97GIVSTFVNSGDD351 pKa = 3.32VATVVRR357 pKa = 11.84VVATMDD363 pKa = 3.14AMNGEE368 pKa = 4.57GTVSAVSDD376 pKa = 3.72VLTISTGLPDD386 pKa = 3.58QNSISLSVEE395 pKa = 3.81GTSVMIEE402 pKa = 3.63DD403 pKa = 4.09AMRR406 pKa = 11.84VDD408 pKa = 5.08GITRR412 pKa = 11.84AITVRR417 pKa = 11.84MADD420 pKa = 3.31KK421 pKa = 10.68FNNPVPDD428 pKa = 3.34GTAAVFTTEE437 pKa = 3.94YY438 pKa = 11.14GSIEE442 pKa = 4.41SACQTTNGACSVTWTSQAPRR462 pKa = 11.84FPTLPEE468 pKa = 3.78NQEE471 pKa = 4.21LVRR474 pKa = 11.84TIYY477 pKa = 10.81DD478 pKa = 3.18ADD480 pKa = 4.04YY481 pKa = 11.25ACPSHH486 pKa = 6.25NANGGPCPDD495 pKa = 3.94DD496 pKa = 4.01LGFIRR501 pKa = 11.84GARR504 pKa = 11.84STILVTAIGEE514 pKa = 4.15EE515 pKa = 4.25SFIDD519 pKa = 3.59ANANGIYY526 pKa = 10.27DD527 pKa = 3.61EE528 pKa = 4.78GEE530 pKa = 3.73RR531 pKa = 11.84FANLTEE537 pKa = 4.7AFLDD541 pKa = 3.99NNWNGFYY548 pKa = 10.9DD549 pKa = 3.84PARR552 pKa = 11.84TYY554 pKa = 10.81CVNNPTLRR562 pKa = 11.84EE563 pKa = 3.9CRR565 pKa = 11.84AGSEE569 pKa = 4.61EE570 pKa = 4.94IFTDD574 pKa = 4.36FNNNGVFDD582 pKa = 5.21ANGDD586 pKa = 3.89DD587 pKa = 4.23PANGYY592 pKa = 9.15PDD594 pKa = 3.69EE595 pKa = 4.88GVTAVYY601 pKa = 10.6NGLLCPPEE609 pKa = 5.79GDD611 pKa = 3.2GVYY614 pKa = 10.44CSRR617 pKa = 11.84EE618 pKa = 4.03LVNVRR623 pKa = 11.84DD624 pKa = 4.26DD625 pKa = 3.83LQIILSDD632 pKa = 3.64NPAWDD637 pKa = 3.48VLVVDD642 pKa = 4.06NRR644 pKa = 11.84GNAVTDD650 pKa = 3.38TAFNGGEE657 pKa = 4.05YY658 pKa = 9.73QVYY661 pKa = 10.23VSDD664 pKa = 4.17TFNNAPIAGTTISVTSSVCEE684 pKa = 3.98VNLSGNTEE692 pKa = 3.99VPNAILPGAFGVSLTQGGSVTYY714 pKa = 10.26ASCRR718 pKa = 11.84DD719 pKa = 3.39EE720 pKa = 4.42EE721 pKa = 4.55RR722 pKa = 11.84PSNTSEE728 pKa = 3.7FQINVGSEE736 pKa = 3.96YY737 pKa = 11.03NEE739 pKa = 4.12TFSCVIQVIDD749 pKa = 4.75DD750 pKa = 4.32ASDD753 pKa = 3.61CPEE756 pKa = 4.35PEE758 pKa = 4.57GGGGGDD764 pKa = 4.45DD765 pKa = 3.76GLSNGGG771 pKa = 3.49
Molecular weight: 79.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165UE41|A0A165UE41_9GAMM Dehydratase OS=Halioglobus sp. HI00S01 OX=1822214 GN=A3709_14885 PE=4 SV=1
MM1 pKa = 7.3VLTACLLVSKK11 pKa = 9.88VNRR14 pKa = 11.84ATRR17 pKa = 11.84AHH19 pKa = 5.59QVQPEE24 pKa = 4.13QQVQPEE30 pKa = 3.93QRR32 pKa = 11.84EE33 pKa = 3.9QRR35 pKa = 11.84EE36 pKa = 4.19QQVLLVPKK44 pKa = 9.33EE45 pKa = 3.95KK46 pKa = 10.39RR47 pKa = 11.84ATRR50 pKa = 11.84VPPAPLAHH58 pKa = 6.43APYY61 pKa = 8.84RR62 pKa = 11.84AKK64 pKa = 10.64RR65 pKa = 11.84PFHH68 pKa = 6.47VVTHH72 pKa = 6.52RR73 pKa = 11.84CQYY76 pKa = 10.08PVFVLILQPFSS87 pKa = 3.34
MM1 pKa = 7.3VLTACLLVSKK11 pKa = 9.88VNRR14 pKa = 11.84ATRR17 pKa = 11.84AHH19 pKa = 5.59QVQPEE24 pKa = 4.13QQVQPEE30 pKa = 3.93QRR32 pKa = 11.84EE33 pKa = 3.9QRR35 pKa = 11.84EE36 pKa = 4.19QQVLLVPKK44 pKa = 9.33EE45 pKa = 3.95KK46 pKa = 10.39RR47 pKa = 11.84ATRR50 pKa = 11.84VPPAPLAHH58 pKa = 6.43APYY61 pKa = 8.84RR62 pKa = 11.84AKK64 pKa = 10.64RR65 pKa = 11.84PFHH68 pKa = 6.47VVTHH72 pKa = 6.52RR73 pKa = 11.84CQYY76 pKa = 10.08PVFVLILQPFSS87 pKa = 3.34
Molecular weight: 10.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1321789 |
38 |
3378 |
325.0 |
35.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.661 ± 0.046 | 1.031 ± 0.014 |
6.166 ± 0.028 | 6.593 ± 0.035 |
3.676 ± 0.023 | 7.933 ± 0.038 |
2.185 ± 0.02 | 5.099 ± 0.028 |
3.212 ± 0.028 | 10.284 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.575 ± 0.018 | 3.257 ± 0.021 |
4.635 ± 0.024 | 3.958 ± 0.022 |
6.071 ± 0.029 | 6.028 ± 0.03 |
5.188 ± 0.023 | 7.173 ± 0.03 |
1.393 ± 0.016 | 2.873 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
