
Planococcus sp. Y42
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Planococcus
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
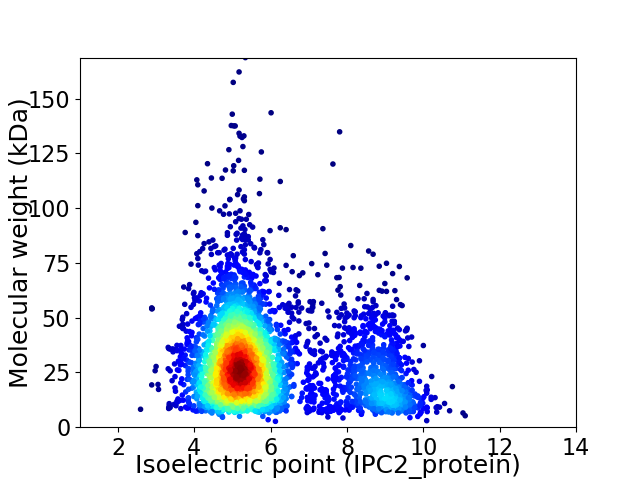
Virtual 2D-PAGE plot for 3921 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q2L493|A0A1Q2L493_9BACL Uncharacterized protein OS=Planococcus sp. Y42 OX=2213202 GN=B0X71_18840 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.62KK3 pKa = 10.49KK4 pKa = 10.17MNLFLLLSTAIMMVGCTDD22 pKa = 3.5AEE24 pKa = 4.21EE25 pKa = 4.3SAVAGEE31 pKa = 4.27EE32 pKa = 4.27TVSQEE37 pKa = 3.74VAEE40 pKa = 4.81AEE42 pKa = 4.09TEE44 pKa = 4.37EE45 pKa = 4.48ISEE48 pKa = 4.15QAEE51 pKa = 4.0TDD53 pKa = 3.48TAVADD58 pKa = 3.94EE59 pKa = 4.93TEE61 pKa = 4.6TEE63 pKa = 3.95ASPEE67 pKa = 4.0EE68 pKa = 4.29SSEE71 pKa = 4.36AEE73 pKa = 3.85AAPSDD78 pKa = 4.39VEE80 pKa = 4.32SAPAQPEE87 pKa = 3.91EE88 pKa = 4.35APFPGYY94 pKa = 10.39EE95 pKa = 4.68LITVDD100 pKa = 5.04GGDD103 pKa = 3.07LSGYY107 pKa = 9.69RR108 pKa = 11.84EE109 pKa = 4.27PNVVVDD115 pKa = 3.08VGYY118 pKa = 10.95GEE120 pKa = 4.04RR121 pKa = 11.84EE122 pKa = 3.6YY123 pKa = 10.44WAFTNEE129 pKa = 3.58YY130 pKa = 8.34GQLVRR135 pKa = 11.84VVADD139 pKa = 4.42EE140 pKa = 6.08IIPQDD145 pKa = 4.07DD146 pKa = 3.47NSEE149 pKa = 4.07PVTADD154 pKa = 2.51GRR156 pKa = 11.84YY157 pKa = 9.03YY158 pKa = 9.98PDD160 pKa = 3.36EE161 pKa = 4.51AKK163 pKa = 11.02VPGVEE168 pKa = 4.89SDD170 pKa = 3.86VLDD173 pKa = 4.46EE174 pKa = 4.33GHH176 pKa = 7.0VIADD180 pKa = 3.72SLGGVANAYY189 pKa = 10.48NITPQDD195 pKa = 3.29STLNRR200 pKa = 11.84HH201 pKa = 5.53GDD203 pKa = 3.34QAYY206 pKa = 8.73MEE208 pKa = 4.63DD209 pKa = 4.09VIRR212 pKa = 11.84DD213 pKa = 3.58AGGATHH219 pKa = 7.12FEE221 pKa = 4.53AIITYY226 pKa = 9.59PNTEE230 pKa = 3.85TQIPSRR236 pKa = 11.84YY237 pKa = 8.33QYY239 pKa = 10.59TYY241 pKa = 9.06TVMGNEE247 pKa = 4.51VVDD250 pKa = 3.48TFNNVNPDD258 pKa = 3.54EE259 pKa = 4.54VNEE262 pKa = 4.18SLGLTEE268 pKa = 5.35NEE270 pKa = 4.05PAEE273 pKa = 4.35PADD276 pKa = 3.58TDD278 pKa = 3.78TAHH281 pKa = 7.23DD282 pKa = 4.03VSSVDD287 pKa = 3.51TNGNGQVTIKK297 pKa = 9.56EE298 pKa = 3.92AKK300 pKa = 9.66AAGFSMPIMSDD311 pKa = 2.56HH312 pKa = 6.56WLYY315 pKa = 10.53PYY317 pKa = 9.86MQDD320 pKa = 3.21NDD322 pKa = 3.64KK323 pKa = 11.57DD324 pKa = 4.13GMVGEE329 pKa = 4.54
MM1 pKa = 7.37KK2 pKa = 10.62KK3 pKa = 10.49KK4 pKa = 10.17MNLFLLLSTAIMMVGCTDD22 pKa = 3.5AEE24 pKa = 4.21EE25 pKa = 4.3SAVAGEE31 pKa = 4.27EE32 pKa = 4.27TVSQEE37 pKa = 3.74VAEE40 pKa = 4.81AEE42 pKa = 4.09TEE44 pKa = 4.37EE45 pKa = 4.48ISEE48 pKa = 4.15QAEE51 pKa = 4.0TDD53 pKa = 3.48TAVADD58 pKa = 3.94EE59 pKa = 4.93TEE61 pKa = 4.6TEE63 pKa = 3.95ASPEE67 pKa = 4.0EE68 pKa = 4.29SSEE71 pKa = 4.36AEE73 pKa = 3.85AAPSDD78 pKa = 4.39VEE80 pKa = 4.32SAPAQPEE87 pKa = 3.91EE88 pKa = 4.35APFPGYY94 pKa = 10.39EE95 pKa = 4.68LITVDD100 pKa = 5.04GGDD103 pKa = 3.07LSGYY107 pKa = 9.69RR108 pKa = 11.84EE109 pKa = 4.27PNVVVDD115 pKa = 3.08VGYY118 pKa = 10.95GEE120 pKa = 4.04RR121 pKa = 11.84EE122 pKa = 3.6YY123 pKa = 10.44WAFTNEE129 pKa = 3.58YY130 pKa = 8.34GQLVRR135 pKa = 11.84VVADD139 pKa = 4.42EE140 pKa = 6.08IIPQDD145 pKa = 4.07DD146 pKa = 3.47NSEE149 pKa = 4.07PVTADD154 pKa = 2.51GRR156 pKa = 11.84YY157 pKa = 9.03YY158 pKa = 9.98PDD160 pKa = 3.36EE161 pKa = 4.51AKK163 pKa = 11.02VPGVEE168 pKa = 4.89SDD170 pKa = 3.86VLDD173 pKa = 4.46EE174 pKa = 4.33GHH176 pKa = 7.0VIADD180 pKa = 3.72SLGGVANAYY189 pKa = 10.48NITPQDD195 pKa = 3.29STLNRR200 pKa = 11.84HH201 pKa = 5.53GDD203 pKa = 3.34QAYY206 pKa = 8.73MEE208 pKa = 4.63DD209 pKa = 4.09VIRR212 pKa = 11.84DD213 pKa = 3.58AGGATHH219 pKa = 7.12FEE221 pKa = 4.53AIITYY226 pKa = 9.59PNTEE230 pKa = 3.85TQIPSRR236 pKa = 11.84YY237 pKa = 8.33QYY239 pKa = 10.59TYY241 pKa = 9.06TVMGNEE247 pKa = 4.51VVDD250 pKa = 3.48TFNNVNPDD258 pKa = 3.54EE259 pKa = 4.54VNEE262 pKa = 4.18SLGLTEE268 pKa = 5.35NEE270 pKa = 4.05PAEE273 pKa = 4.35PADD276 pKa = 3.58TDD278 pKa = 3.78TAHH281 pKa = 7.23DD282 pKa = 4.03VSSVDD287 pKa = 3.51TNGNGQVTIKK297 pKa = 9.56EE298 pKa = 3.92AKK300 pKa = 9.66AAGFSMPIMSDD311 pKa = 2.56HH312 pKa = 6.56WLYY315 pKa = 10.53PYY317 pKa = 9.86MQDD320 pKa = 3.21NDD322 pKa = 3.64KK323 pKa = 11.57DD324 pKa = 4.13GMVGEE329 pKa = 4.54
Molecular weight: 35.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q2KVC1|A0A1Q2KVC1_9BACL Uncharacterized protein OS=Planococcus sp. Y42 OX=2213202 GN=B0X71_02645 PE=4 SV=1
MM1 pKa = 7.34FFHH4 pKa = 6.8NRR6 pKa = 11.84TASAAKK12 pKa = 9.62ARR14 pKa = 11.84RR15 pKa = 11.84IKK17 pKa = 9.97TSAPAPLRR25 pKa = 11.84LLVANSSGKK34 pKa = 10.16AVAAAVRR41 pKa = 11.84WLPPFHH47 pKa = 7.1SRR49 pKa = 11.84QLKK52 pKa = 6.71MTTRR56 pKa = 11.84QNAA59 pKa = 3.02
MM1 pKa = 7.34FFHH4 pKa = 6.8NRR6 pKa = 11.84TASAAKK12 pKa = 9.62ARR14 pKa = 11.84RR15 pKa = 11.84IKK17 pKa = 9.97TSAPAPLRR25 pKa = 11.84LLVANSSGKK34 pKa = 10.16AVAAAVRR41 pKa = 11.84WLPPFHH47 pKa = 7.1SRR49 pKa = 11.84QLKK52 pKa = 6.71MTTRR56 pKa = 11.84QNAA59 pKa = 3.02
Molecular weight: 6.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1117653 |
25 |
1528 |
285.0 |
31.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.656 ± 0.045 | 0.62 ± 0.01 |
5.32 ± 0.031 | 8.04 ± 0.05 |
4.481 ± 0.026 | 7.267 ± 0.037 |
2.05 ± 0.018 | 6.715 ± 0.036 |
5.463 ± 0.042 | 9.896 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.682 ± 0.018 | 3.657 ± 0.026 |
3.933 ± 0.027 | 3.771 ± 0.025 |
4.892 ± 0.03 | 5.598 ± 0.026 |
5.557 ± 0.025 | 7.088 ± 0.036 |
1.062 ± 0.014 | 3.253 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
