
Odocoileus virginianus papillomavirus 1 (DPV) (Deer papillomavirus)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Deltapapillomavirus; Deltapapillomavirus 2
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
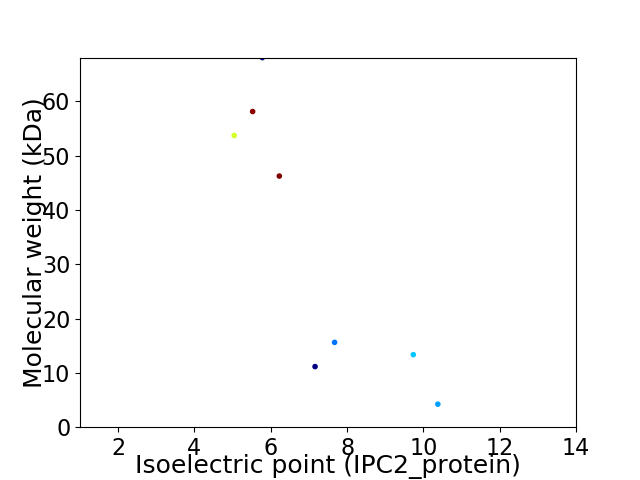
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03117|VE1_OVPVD Replication protein E1 OS=Odocoileus virginianus papillomavirus 1 OX=2772504 GN=E1 PE=3 SV=2
MM1 pKa = 7.74PPLKK5 pKa = 9.91RR6 pKa = 11.84VKK8 pKa = 10.17RR9 pKa = 11.84ANPYY13 pKa = 10.57DD14 pKa = 3.99LYY16 pKa = 10.14RR17 pKa = 11.84TCKK20 pKa = 9.47RR21 pKa = 11.84WKK23 pKa = 9.5CPLMSFLKK31 pKa = 10.98VEE33 pKa = 4.52GKK35 pKa = 8.16TVADD39 pKa = 5.18KK40 pKa = 9.86YY41 pKa = 11.18CSMGSMGVYY50 pKa = 8.77WRR52 pKa = 11.84LALHH56 pKa = 6.52GSGRR60 pKa = 11.84PTQGGYY66 pKa = 10.17VPLRR70 pKa = 11.84GGGSSTSLSSRR81 pKa = 11.84GSGSSTSISRR91 pKa = 11.84PFAGGIPLEE100 pKa = 4.18TLEE103 pKa = 4.42TVGAFRR109 pKa = 11.84PGIIEE114 pKa = 3.95EE115 pKa = 4.24VAPTLEE121 pKa = 4.42GVLPDD126 pKa = 3.8APAVVTPEE134 pKa = 4.29AVPVDD139 pKa = 3.71QGLSGLDD146 pKa = 3.2VARR149 pKa = 11.84EE150 pKa = 4.1VTQEE154 pKa = 3.85SLITFLQPEE163 pKa = 4.94GPDD166 pKa = 3.93DD167 pKa = 3.69IAVLEE172 pKa = 4.23LRR174 pKa = 11.84PTEE177 pKa = 4.11HH178 pKa = 7.18DD179 pKa = 3.34QTHH182 pKa = 7.13LISTSTHH189 pKa = 6.26PNPLFHH195 pKa = 7.52APIQQSSIIAEE206 pKa = 4.2TSGSEE211 pKa = 4.03NIFVGGGGVGSTTGEE226 pKa = 4.18EE227 pKa = 3.98IEE229 pKa = 4.48LTLFGQPKK237 pKa = 8.73TSTPEE242 pKa = 3.79GPINRR247 pKa = 11.84GRR249 pKa = 11.84GIFNWFNRR257 pKa = 11.84TYY259 pKa = 8.2YY260 pKa = 8.89TQVPVEE266 pKa = 4.88DD267 pKa = 4.44PDD269 pKa = 4.14EE270 pKa = 4.1IAAAGSYY277 pKa = 9.78VFEE280 pKa = 4.31NALYY284 pKa = 10.27DD285 pKa = 3.81SKK287 pKa = 11.41AYY289 pKa = 9.72KK290 pKa = 10.18HH291 pKa = 6.08EE292 pKa = 3.99QQPWLSRR299 pKa = 11.84PQDD302 pKa = 3.38APEE305 pKa = 4.62FDD307 pKa = 3.81FQDD310 pKa = 3.69AVRR313 pKa = 11.84LLQGPSGRR321 pKa = 11.84VGWSRR326 pKa = 11.84IIRR329 pKa = 11.84PTSIGTRR336 pKa = 11.84SGVRR340 pKa = 11.84VGPLYY345 pKa = 10.53HH346 pKa = 6.78LRR348 pKa = 11.84QSFSTIDD355 pKa = 3.46EE356 pKa = 4.63PEE358 pKa = 4.32TIEE361 pKa = 5.52LIPSTVDD368 pKa = 3.0EE369 pKa = 4.78EE370 pKa = 4.5EE371 pKa = 4.36VLTGVPEE378 pKa = 4.27SAEE381 pKa = 4.24GPDD384 pKa = 4.57AEE386 pKa = 4.87YY387 pKa = 11.31SDD389 pKa = 5.7IDD391 pKa = 3.89LQSIGSDD398 pKa = 3.35EE399 pKa = 4.36PLLGTGIIYY408 pKa = 9.81PLVGGGQIFLCMHH421 pKa = 7.58RR422 pKa = 11.84APVGWSSGTYY432 pKa = 9.52INHH435 pKa = 6.77EE436 pKa = 4.14GQSRR440 pKa = 11.84DD441 pKa = 3.37DD442 pKa = 3.7GEE444 pKa = 4.34YY445 pKa = 10.93VIDD448 pKa = 5.49NGGQSNITPTVVIDD462 pKa = 3.74GSIALSLEE470 pKa = 4.25YY471 pKa = 10.11FRR473 pKa = 11.84HH474 pKa = 6.0YY475 pKa = 10.47YY476 pKa = 10.03LHH478 pKa = 7.25PSLLRR483 pKa = 11.84RR484 pKa = 11.84KK485 pKa = 9.7RR486 pKa = 11.84KK487 pKa = 9.64RR488 pKa = 11.84NPIFII493 pKa = 4.84
MM1 pKa = 7.74PPLKK5 pKa = 9.91RR6 pKa = 11.84VKK8 pKa = 10.17RR9 pKa = 11.84ANPYY13 pKa = 10.57DD14 pKa = 3.99LYY16 pKa = 10.14RR17 pKa = 11.84TCKK20 pKa = 9.47RR21 pKa = 11.84WKK23 pKa = 9.5CPLMSFLKK31 pKa = 10.98VEE33 pKa = 4.52GKK35 pKa = 8.16TVADD39 pKa = 5.18KK40 pKa = 9.86YY41 pKa = 11.18CSMGSMGVYY50 pKa = 8.77WRR52 pKa = 11.84LALHH56 pKa = 6.52GSGRR60 pKa = 11.84PTQGGYY66 pKa = 10.17VPLRR70 pKa = 11.84GGGSSTSLSSRR81 pKa = 11.84GSGSSTSISRR91 pKa = 11.84PFAGGIPLEE100 pKa = 4.18TLEE103 pKa = 4.42TVGAFRR109 pKa = 11.84PGIIEE114 pKa = 3.95EE115 pKa = 4.24VAPTLEE121 pKa = 4.42GVLPDD126 pKa = 3.8APAVVTPEE134 pKa = 4.29AVPVDD139 pKa = 3.71QGLSGLDD146 pKa = 3.2VARR149 pKa = 11.84EE150 pKa = 4.1VTQEE154 pKa = 3.85SLITFLQPEE163 pKa = 4.94GPDD166 pKa = 3.93DD167 pKa = 3.69IAVLEE172 pKa = 4.23LRR174 pKa = 11.84PTEE177 pKa = 4.11HH178 pKa = 7.18DD179 pKa = 3.34QTHH182 pKa = 7.13LISTSTHH189 pKa = 6.26PNPLFHH195 pKa = 7.52APIQQSSIIAEE206 pKa = 4.2TSGSEE211 pKa = 4.03NIFVGGGGVGSTTGEE226 pKa = 4.18EE227 pKa = 3.98IEE229 pKa = 4.48LTLFGQPKK237 pKa = 8.73TSTPEE242 pKa = 3.79GPINRR247 pKa = 11.84GRR249 pKa = 11.84GIFNWFNRR257 pKa = 11.84TYY259 pKa = 8.2YY260 pKa = 8.89TQVPVEE266 pKa = 4.88DD267 pKa = 4.44PDD269 pKa = 4.14EE270 pKa = 4.1IAAAGSYY277 pKa = 9.78VFEE280 pKa = 4.31NALYY284 pKa = 10.27DD285 pKa = 3.81SKK287 pKa = 11.41AYY289 pKa = 9.72KK290 pKa = 10.18HH291 pKa = 6.08EE292 pKa = 3.99QQPWLSRR299 pKa = 11.84PQDD302 pKa = 3.38APEE305 pKa = 4.62FDD307 pKa = 3.81FQDD310 pKa = 3.69AVRR313 pKa = 11.84LLQGPSGRR321 pKa = 11.84VGWSRR326 pKa = 11.84IIRR329 pKa = 11.84PTSIGTRR336 pKa = 11.84SGVRR340 pKa = 11.84VGPLYY345 pKa = 10.53HH346 pKa = 6.78LRR348 pKa = 11.84QSFSTIDD355 pKa = 3.46EE356 pKa = 4.63PEE358 pKa = 4.32TIEE361 pKa = 5.52LIPSTVDD368 pKa = 3.0EE369 pKa = 4.78EE370 pKa = 4.5EE371 pKa = 4.36VLTGVPEE378 pKa = 4.27SAEE381 pKa = 4.24GPDD384 pKa = 4.57AEE386 pKa = 4.87YY387 pKa = 11.31SDD389 pKa = 5.7IDD391 pKa = 3.89LQSIGSDD398 pKa = 3.35EE399 pKa = 4.36PLLGTGIIYY408 pKa = 9.81PLVGGGQIFLCMHH421 pKa = 7.58RR422 pKa = 11.84APVGWSSGTYY432 pKa = 9.52INHH435 pKa = 6.77EE436 pKa = 4.14GQSRR440 pKa = 11.84DD441 pKa = 3.37DD442 pKa = 3.7GEE444 pKa = 4.34YY445 pKa = 10.93VIDD448 pKa = 5.49NGGQSNITPTVVIDD462 pKa = 3.74GSIALSLEE470 pKa = 4.25YY471 pKa = 10.11FRR473 pKa = 11.84HH474 pKa = 6.0YY475 pKa = 10.47YY476 pKa = 10.03LHH478 pKa = 7.25PSLLRR483 pKa = 11.84RR484 pKa = 11.84KK485 pKa = 9.7RR486 pKa = 11.84KK487 pKa = 9.64RR488 pKa = 11.84NPIFII493 pKa = 4.84
Molecular weight: 53.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P06925|VE4_OVPVD Probable protein E4 OS=Odocoileus virginianus papillomavirus 1 OX=2772504 GN=E4 PE=4 SV=1
GG1 pKa = 5.97ITIVLITHH9 pKa = 6.88ILRR12 pKa = 11.84PPTLEE17 pKa = 4.86RR18 pKa = 11.84PSKK21 pKa = 11.04DD22 pKa = 3.0CGTPGAVKK30 pKa = 10.12EE31 pKa = 4.38ADD33 pKa = 3.46PPTRR37 pKa = 11.84PTAPCFTLLLEE48 pKa = 4.5ATPFTVPSEE57 pKa = 3.98LAKK60 pKa = 10.34TGVGPLTARR69 pKa = 11.84LPTAHH74 pKa = 7.03HH75 pKa = 6.43SPRR78 pKa = 11.84GVAWAPIPPPRR89 pKa = 11.84CRR91 pKa = 11.84ARR93 pKa = 11.84YY94 pKa = 9.02RR95 pKa = 11.84RR96 pKa = 11.84TPGAYY101 pKa = 10.25LYY103 pKa = 7.52PTVLDD108 pKa = 3.46EE109 pKa = 4.21GRR111 pKa = 11.84RR112 pKa = 11.84ITRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84QINTTT122 pKa = 3.32
GG1 pKa = 5.97ITIVLITHH9 pKa = 6.88ILRR12 pKa = 11.84PPTLEE17 pKa = 4.86RR18 pKa = 11.84PSKK21 pKa = 11.04DD22 pKa = 3.0CGTPGAVKK30 pKa = 10.12EE31 pKa = 4.38ADD33 pKa = 3.46PPTRR37 pKa = 11.84PTAPCFTLLLEE48 pKa = 4.5ATPFTVPSEE57 pKa = 3.98LAKK60 pKa = 10.34TGVGPLTARR69 pKa = 11.84LPTAHH74 pKa = 7.03HH75 pKa = 6.43SPRR78 pKa = 11.84GVAWAPIPPPRR89 pKa = 11.84CRR91 pKa = 11.84ARR93 pKa = 11.84YY94 pKa = 9.02RR95 pKa = 11.84RR96 pKa = 11.84TPGAYY101 pKa = 10.25LYY103 pKa = 7.52PTVLDD108 pKa = 3.46EE109 pKa = 4.21GRR111 pKa = 11.84RR112 pKa = 11.84ITRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84QINTTT122 pKa = 3.32
Molecular weight: 13.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2429 |
35 |
613 |
303.6 |
33.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.175 ± 0.692 | 2.758 ± 0.744 |
5.105 ± 0.297 | 6.093 ± 0.318 |
3.541 ± 0.469 | 7.452 ± 0.876 |
2.594 ± 0.297 | 4.199 ± 0.62 |
4.446 ± 0.624 | 8.687 ± 0.383 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.811 ± 0.354 | 3.746 ± 0.571 |
7.369 ± 1.13 | 3.788 ± 0.262 |
6.175 ± 0.893 | 7.657 ± 0.706 |
7.122 ± 0.652 | 6.093 ± 0.303 |
1.688 ± 0.315 | 3.499 ± 0.5 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
