
Phytoactinopolyspora alkaliphila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jiangellales; Jiangellaceae; Phytoactinopolyspora
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
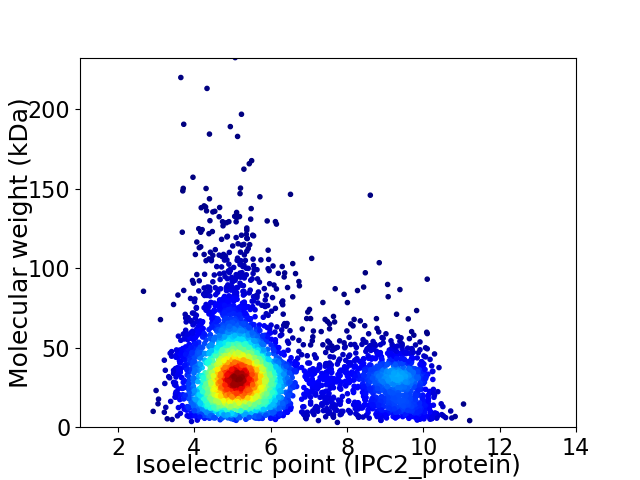
Virtual 2D-PAGE plot for 4852 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N9YTC5|A0A6N9YTC5_9ACTN Cysteine desulfurase OS=Phytoactinopolyspora alkaliphila OX=1783498 GN=G1H11_23615 PE=3 SV=1
MM1 pKa = 5.6TTKK4 pKa = 10.42RR5 pKa = 11.84RR6 pKa = 11.84GAAFLAVAAASALVLAACGGDD27 pKa = 4.18DD28 pKa = 5.22ADD30 pKa = 4.58EE31 pKa = 4.64TPADD35 pKa = 4.35DD36 pKa = 5.47DD37 pKa = 4.11GGAAEE42 pKa = 4.74EE43 pKa = 4.7QPDD46 pKa = 4.36PDD48 pKa = 4.02TGEE51 pKa = 4.37GPSDD55 pKa = 3.79VEE57 pKa = 3.92VVYY60 pKa = 10.41AAEE63 pKa = 4.28QEE65 pKa = 4.3FTNYY69 pKa = 10.61ANNTSEE75 pKa = 4.87GNLLANTHH83 pKa = 5.66VLNQVKK89 pKa = 10.4GGFWNYY95 pKa = 10.35GPDD98 pKa = 3.58GLVVPDD104 pKa = 4.71EE105 pKa = 4.31EE106 pKa = 4.64FGTYY110 pKa = 10.69DD111 pKa = 4.63LVSDD115 pKa = 4.69DD116 pKa = 4.0PQTIEE121 pKa = 5.07FNINPDD127 pKa = 3.79AVWSDD132 pKa = 4.06GEE134 pKa = 5.26PIDD137 pKa = 5.81CDD139 pKa = 4.29DD140 pKa = 5.77FLLGWAAQSGYY151 pKa = 9.91YY152 pKa = 8.26EE153 pKa = 4.98HH154 pKa = 7.15PTEE157 pKa = 4.24TTADD161 pKa = 3.91GEE163 pKa = 4.33PATLFSTAGTTGYY176 pKa = 11.04ADD178 pKa = 3.39WVKK181 pKa = 10.48PDD183 pKa = 4.7CEE185 pKa = 4.96PGDD188 pKa = 3.47KK189 pKa = 10.43TITVEE194 pKa = 4.16YY195 pKa = 8.09EE196 pKa = 3.97TVNASWVLYY205 pKa = 8.96TAVDD209 pKa = 3.97MPAHH213 pKa = 4.91VVARR217 pKa = 11.84EE218 pKa = 3.75GGMASGDD225 pKa = 3.75EE226 pKa = 4.5LIEE229 pKa = 4.85AIRR232 pKa = 11.84NDD234 pKa = 5.23DD235 pKa = 3.7IDD237 pKa = 4.61ALLPAAEE244 pKa = 4.79FYY246 pKa = 9.79NTGFVMSPGEE256 pKa = 4.19LLPEE260 pKa = 4.48EE261 pKa = 5.25IIPSSGRR268 pKa = 11.84YY269 pKa = 9.37KK270 pKa = 10.2LTDD273 pKa = 3.12WQAGQSITMEE283 pKa = 4.03YY284 pKa = 10.07NEE286 pKa = 5.19NYY288 pKa = 9.63WGTPPAARR296 pKa = 11.84TIIFRR301 pKa = 11.84FLDD304 pKa = 3.73ANQQAQALQNQEE316 pKa = 3.74IDD318 pKa = 3.62IMDD321 pKa = 4.59PQPSTDD327 pKa = 4.18LVNQLEE333 pKa = 4.41GMAGVEE339 pKa = 4.03VHH341 pKa = 6.12VDD343 pKa = 3.19EE344 pKa = 5.09SFTYY348 pKa = 10.16EE349 pKa = 3.91HH350 pKa = 6.92VDD352 pKa = 3.64FNLRR356 pKa = 11.84GDD358 pKa = 4.16HH359 pKa = 6.47YY360 pKa = 11.58FNDD363 pKa = 3.67PNLRR367 pKa = 11.84RR368 pKa = 11.84SFGLCLPRR376 pKa = 11.84QLMVEE381 pKa = 4.11NLIHH385 pKa = 6.19PQNPDD390 pKa = 3.24AEE392 pKa = 4.53VLNARR397 pKa = 11.84LTYY400 pKa = 10.49SFQPDD405 pKa = 3.26YY406 pKa = 11.48DD407 pKa = 3.75FQIDD411 pKa = 3.77GNGWEE416 pKa = 5.11DD417 pKa = 4.21FAEE420 pKa = 3.92QDD422 pKa = 2.75IDD424 pKa = 3.4EE425 pKa = 4.38SRR427 pKa = 11.84RR428 pKa = 11.84LLEE431 pKa = 4.18EE432 pKa = 5.14ADD434 pKa = 3.61MVGTEE439 pKa = 3.69VRR441 pKa = 11.84IVYY444 pKa = 9.09TEE446 pKa = 3.88ANPRR450 pKa = 11.84RR451 pKa = 11.84VDD453 pKa = 3.56QVALIRR459 pKa = 11.84DD460 pKa = 3.85ACNQAGWDD468 pKa = 3.78VVDD471 pKa = 5.33AGSADD476 pKa = 3.62PFGVEE481 pKa = 4.05IPDD484 pKa = 3.8GNFDD488 pKa = 3.23VAMYY492 pKa = 10.36AWIGSGYY499 pKa = 9.93VAGTASTFMTPSSCTPEE516 pKa = 3.9GTGNNSQCYY525 pKa = 8.78SSEE528 pKa = 4.19VVDD531 pKa = 4.04DD532 pKa = 4.98LYY534 pKa = 11.39RR535 pKa = 11.84EE536 pKa = 4.19LLQEE540 pKa = 4.02VDD542 pKa = 2.97VDD544 pKa = 3.84AQRR547 pKa = 11.84EE548 pKa = 4.18LVKK551 pKa = 10.79QIEE554 pKa = 4.45TQLWADD560 pKa = 4.51LPTIPLFTHH569 pKa = 7.28PYY571 pKa = 7.97LLAWAEE577 pKa = 4.14DD578 pKa = 4.07VEE580 pKa = 5.52GITPNPTQQGVTSTKK595 pKa = 10.46HH596 pKa = 3.81EE597 pKa = 4.31WNRR600 pKa = 11.84ALL602 pKa = 4.83
MM1 pKa = 5.6TTKK4 pKa = 10.42RR5 pKa = 11.84RR6 pKa = 11.84GAAFLAVAAASALVLAACGGDD27 pKa = 4.18DD28 pKa = 5.22ADD30 pKa = 4.58EE31 pKa = 4.64TPADD35 pKa = 4.35DD36 pKa = 5.47DD37 pKa = 4.11GGAAEE42 pKa = 4.74EE43 pKa = 4.7QPDD46 pKa = 4.36PDD48 pKa = 4.02TGEE51 pKa = 4.37GPSDD55 pKa = 3.79VEE57 pKa = 3.92VVYY60 pKa = 10.41AAEE63 pKa = 4.28QEE65 pKa = 4.3FTNYY69 pKa = 10.61ANNTSEE75 pKa = 4.87GNLLANTHH83 pKa = 5.66VLNQVKK89 pKa = 10.4GGFWNYY95 pKa = 10.35GPDD98 pKa = 3.58GLVVPDD104 pKa = 4.71EE105 pKa = 4.31EE106 pKa = 4.64FGTYY110 pKa = 10.69DD111 pKa = 4.63LVSDD115 pKa = 4.69DD116 pKa = 4.0PQTIEE121 pKa = 5.07FNINPDD127 pKa = 3.79AVWSDD132 pKa = 4.06GEE134 pKa = 5.26PIDD137 pKa = 5.81CDD139 pKa = 4.29DD140 pKa = 5.77FLLGWAAQSGYY151 pKa = 9.91YY152 pKa = 8.26EE153 pKa = 4.98HH154 pKa = 7.15PTEE157 pKa = 4.24TTADD161 pKa = 3.91GEE163 pKa = 4.33PATLFSTAGTTGYY176 pKa = 11.04ADD178 pKa = 3.39WVKK181 pKa = 10.48PDD183 pKa = 4.7CEE185 pKa = 4.96PGDD188 pKa = 3.47KK189 pKa = 10.43TITVEE194 pKa = 4.16YY195 pKa = 8.09EE196 pKa = 3.97TVNASWVLYY205 pKa = 8.96TAVDD209 pKa = 3.97MPAHH213 pKa = 4.91VVARR217 pKa = 11.84EE218 pKa = 3.75GGMASGDD225 pKa = 3.75EE226 pKa = 4.5LIEE229 pKa = 4.85AIRR232 pKa = 11.84NDD234 pKa = 5.23DD235 pKa = 3.7IDD237 pKa = 4.61ALLPAAEE244 pKa = 4.79FYY246 pKa = 9.79NTGFVMSPGEE256 pKa = 4.19LLPEE260 pKa = 4.48EE261 pKa = 5.25IIPSSGRR268 pKa = 11.84YY269 pKa = 9.37KK270 pKa = 10.2LTDD273 pKa = 3.12WQAGQSITMEE283 pKa = 4.03YY284 pKa = 10.07NEE286 pKa = 5.19NYY288 pKa = 9.63WGTPPAARR296 pKa = 11.84TIIFRR301 pKa = 11.84FLDD304 pKa = 3.73ANQQAQALQNQEE316 pKa = 3.74IDD318 pKa = 3.62IMDD321 pKa = 4.59PQPSTDD327 pKa = 4.18LVNQLEE333 pKa = 4.41GMAGVEE339 pKa = 4.03VHH341 pKa = 6.12VDD343 pKa = 3.19EE344 pKa = 5.09SFTYY348 pKa = 10.16EE349 pKa = 3.91HH350 pKa = 6.92VDD352 pKa = 3.64FNLRR356 pKa = 11.84GDD358 pKa = 4.16HH359 pKa = 6.47YY360 pKa = 11.58FNDD363 pKa = 3.67PNLRR367 pKa = 11.84RR368 pKa = 11.84SFGLCLPRR376 pKa = 11.84QLMVEE381 pKa = 4.11NLIHH385 pKa = 6.19PQNPDD390 pKa = 3.24AEE392 pKa = 4.53VLNARR397 pKa = 11.84LTYY400 pKa = 10.49SFQPDD405 pKa = 3.26YY406 pKa = 11.48DD407 pKa = 3.75FQIDD411 pKa = 3.77GNGWEE416 pKa = 5.11DD417 pKa = 4.21FAEE420 pKa = 3.92QDD422 pKa = 2.75IDD424 pKa = 3.4EE425 pKa = 4.38SRR427 pKa = 11.84RR428 pKa = 11.84LLEE431 pKa = 4.18EE432 pKa = 5.14ADD434 pKa = 3.61MVGTEE439 pKa = 3.69VRR441 pKa = 11.84IVYY444 pKa = 9.09TEE446 pKa = 3.88ANPRR450 pKa = 11.84RR451 pKa = 11.84VDD453 pKa = 3.56QVALIRR459 pKa = 11.84DD460 pKa = 3.85ACNQAGWDD468 pKa = 3.78VVDD471 pKa = 5.33AGSADD476 pKa = 3.62PFGVEE481 pKa = 4.05IPDD484 pKa = 3.8GNFDD488 pKa = 3.23VAMYY492 pKa = 10.36AWIGSGYY499 pKa = 9.93VAGTASTFMTPSSCTPEE516 pKa = 3.9GTGNNSQCYY525 pKa = 8.78SSEE528 pKa = 4.19VVDD531 pKa = 4.04DD532 pKa = 4.98LYY534 pKa = 11.39RR535 pKa = 11.84EE536 pKa = 4.19LLQEE540 pKa = 4.02VDD542 pKa = 2.97VDD544 pKa = 3.84AQRR547 pKa = 11.84EE548 pKa = 4.18LVKK551 pKa = 10.79QIEE554 pKa = 4.45TQLWADD560 pKa = 4.51LPTIPLFTHH569 pKa = 7.28PYY571 pKa = 7.97LLAWAEE577 pKa = 4.14DD578 pKa = 4.07VEE580 pKa = 5.52GITPNPTQQGVTSTKK595 pKa = 10.46HH596 pKa = 3.81EE597 pKa = 4.31WNRR600 pKa = 11.84ALL602 pKa = 4.83
Molecular weight: 66.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N9YI09|A0A6N9YI09_9ACTN Dolichyl-phosphate-mannose--protein mannosyltransferase OS=Phytoactinopolyspora alkaliphila OX=1783498 GN=G1H11_04600 PE=3 SV=1
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.64HH17 pKa = 5.77RR18 pKa = 11.84KK19 pKa = 7.99LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84QGKK33 pKa = 8.88
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.64HH17 pKa = 5.77RR18 pKa = 11.84KK19 pKa = 7.99LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84QGKK33 pKa = 8.88
Molecular weight: 4.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1624139 |
29 |
2147 |
334.7 |
35.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.797 ± 0.041 | 0.677 ± 0.009 |
6.62 ± 0.035 | 5.992 ± 0.027 |
2.876 ± 0.021 | 9.113 ± 0.031 |
2.281 ± 0.018 | 3.866 ± 0.023 |
1.622 ± 0.021 | 9.868 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.943 ± 0.013 | 1.912 ± 0.018 |
5.677 ± 0.023 | 2.769 ± 0.016 |
7.802 ± 0.036 | 5.54 ± 0.023 |
5.908 ± 0.024 | 9.067 ± 0.034 |
1.567 ± 0.013 | 2.102 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
