
Pseudorhizobium pelagicum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Pseudorhizobium
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
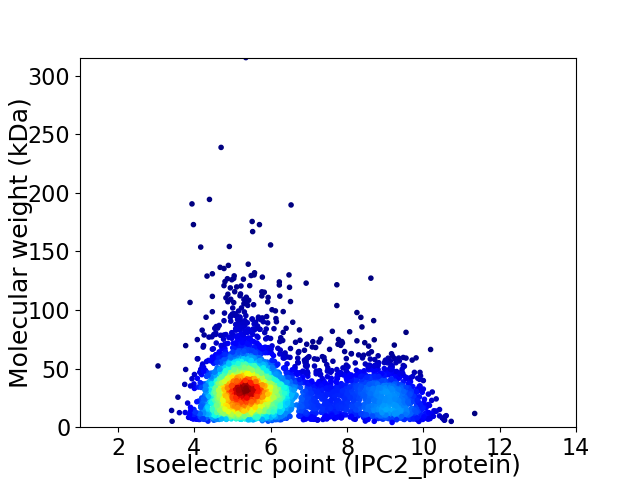
Virtual 2D-PAGE plot for 4417 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A081MES3|A0A081MES3_9RHIZ UPF0060 membrane protein GV67_07810 OS=Pseudorhizobium pelagicum OX=1509405 GN=GV67_07810 PE=3 SV=1
MM1 pKa = 7.56AVLDD5 pKa = 4.33QITEE9 pKa = 4.41EE10 pKa = 3.84ISHH13 pKa = 5.92FVGFFHH19 pKa = 7.65LSLEE23 pKa = 4.28EE24 pKa = 3.72ARR26 pKa = 11.84QRR28 pKa = 11.84EE29 pKa = 4.56SYY31 pKa = 11.42NDD33 pKa = 3.45FSFKK37 pKa = 8.4TTLHH41 pKa = 6.14GVDD44 pKa = 3.23SRR46 pKa = 11.84LVEE49 pKa = 4.28EE50 pKa = 5.07APFAAPHH57 pKa = 6.53ALDD60 pKa = 5.45DD61 pKa = 4.1YY62 pKa = 11.9DD63 pKa = 5.69PILNYY68 pKa = 10.46RR69 pKa = 11.84PVAPIFPAAPAFYY82 pKa = 10.35LPPALLDD89 pKa = 3.5VAPEE93 pKa = 3.89NLRR96 pKa = 11.84VEE98 pKa = 4.73VFAQEE103 pKa = 3.86ASLRR107 pKa = 11.84TPMPFHH113 pKa = 6.72TGGKK117 pKa = 7.48ATAPALEE124 pKa = 4.72PPGSLATYY132 pKa = 8.18AQQHH136 pKa = 5.75IVLSDD141 pKa = 3.35NDD143 pKa = 3.68VFSVGGHH150 pKa = 5.86GFSISGNPVALAEE163 pKa = 4.41LLSMAEE169 pKa = 4.06PVLAVSPLGEE179 pKa = 4.17LEE181 pKa = 4.5APGSAAEE188 pKa = 4.42MIDD191 pKa = 3.68LVEE194 pKa = 4.46TVAEE198 pKa = 4.25SLEE201 pKa = 3.93HH202 pKa = 7.02LIDD205 pKa = 4.02GAGSHH210 pKa = 5.73QVAQASTLSGVYY222 pKa = 9.59VNGVAVEE229 pKa = 3.99QAPSLEE235 pKa = 5.0DD236 pKa = 3.13YY237 pKa = 11.09HH238 pKa = 9.11SFDD241 pKa = 4.16EE242 pKa = 5.43GEE244 pKa = 4.29DD245 pKa = 3.3DD246 pKa = 4.11GAIVEE251 pKa = 4.64AEE253 pKa = 3.76WGGEE257 pKa = 4.14GSQPPPASVTITMGDD272 pKa = 3.18NTLVNDD278 pKa = 4.03VLVKK282 pKa = 10.73NLWTAAKK289 pKa = 8.48VTVVQGDD296 pKa = 4.04HH297 pKa = 6.68VEE299 pKa = 4.31VNAVVQINAVWDD311 pKa = 3.71TDD313 pKa = 4.51SIGSTVAGWGASNSANDD330 pKa = 4.17LFNIASFDD338 pKa = 3.84HH339 pKa = 7.49DD340 pKa = 4.78DD341 pKa = 3.93SAGSQQLAGASRR353 pKa = 11.84PDD355 pKa = 4.3LYY357 pKa = 10.51PSYY360 pKa = 10.74WNVTTVTGDD369 pKa = 3.35LMIVNWIEE377 pKa = 3.59QLIFMSDD384 pKa = 2.98ADD386 pKa = 3.93MGIVSASGAYY396 pKa = 8.37STIISGDD403 pKa = 3.27NTAVNHH409 pKa = 4.6TTITEE414 pKa = 4.14LGFGYY419 pKa = 10.59DD420 pKa = 4.11LIIIGGSVYY429 pKa = 10.48DD430 pKa = 4.16ANIIQQVNVLFDD442 pKa = 3.74NDD444 pKa = 3.97AVTTTDD450 pKa = 3.05GFEE453 pKa = 4.03SAEE456 pKa = 4.17AGTVSASGNLLWNQASIYY474 pKa = 9.68TIGAAEE480 pKa = 4.13RR481 pKa = 11.84FGEE484 pKa = 4.46LSDD487 pKa = 3.97AYY489 pKa = 10.28RR490 pKa = 11.84DD491 pKa = 3.75AVASLQSGSANLSDD505 pKa = 4.57EE506 pKa = 4.3VLHH509 pKa = 7.12DD510 pKa = 3.75SAFAGLAGLRR520 pKa = 11.84VLHH523 pKa = 5.72VQGDD527 pKa = 4.74FINVQYY533 pKa = 10.74IKK535 pKa = 9.15QTSIIGDD542 pKa = 3.4NDD544 pKa = 3.8QILLAMDD551 pKa = 5.22ALTPNNDD558 pKa = 3.52ASWTVQAGGNTLINNAAILDD578 pKa = 4.0LDD580 pKa = 4.34SFGKK584 pKa = 9.18TYY586 pKa = 11.01VGGEE590 pKa = 4.09QYY592 pKa = 10.88SQEE595 pKa = 4.17TLIQANFISSQPEE608 pKa = 4.12LASHH612 pKa = 7.68DD613 pKa = 4.3PGTLATEE620 pKa = 4.18AVLFLDD626 pKa = 4.21DD627 pKa = 4.34TMLEE631 pKa = 4.16ADD633 pKa = 4.37PAPEE637 pKa = 4.81GGSYY641 pKa = 9.21WPADD645 pKa = 3.59SAGGQDD651 pKa = 5.78DD652 pKa = 4.28GLQTLLAA659 pKa = 4.9
MM1 pKa = 7.56AVLDD5 pKa = 4.33QITEE9 pKa = 4.41EE10 pKa = 3.84ISHH13 pKa = 5.92FVGFFHH19 pKa = 7.65LSLEE23 pKa = 4.28EE24 pKa = 3.72ARR26 pKa = 11.84QRR28 pKa = 11.84EE29 pKa = 4.56SYY31 pKa = 11.42NDD33 pKa = 3.45FSFKK37 pKa = 8.4TTLHH41 pKa = 6.14GVDD44 pKa = 3.23SRR46 pKa = 11.84LVEE49 pKa = 4.28EE50 pKa = 5.07APFAAPHH57 pKa = 6.53ALDD60 pKa = 5.45DD61 pKa = 4.1YY62 pKa = 11.9DD63 pKa = 5.69PILNYY68 pKa = 10.46RR69 pKa = 11.84PVAPIFPAAPAFYY82 pKa = 10.35LPPALLDD89 pKa = 3.5VAPEE93 pKa = 3.89NLRR96 pKa = 11.84VEE98 pKa = 4.73VFAQEE103 pKa = 3.86ASLRR107 pKa = 11.84TPMPFHH113 pKa = 6.72TGGKK117 pKa = 7.48ATAPALEE124 pKa = 4.72PPGSLATYY132 pKa = 8.18AQQHH136 pKa = 5.75IVLSDD141 pKa = 3.35NDD143 pKa = 3.68VFSVGGHH150 pKa = 5.86GFSISGNPVALAEE163 pKa = 4.41LLSMAEE169 pKa = 4.06PVLAVSPLGEE179 pKa = 4.17LEE181 pKa = 4.5APGSAAEE188 pKa = 4.42MIDD191 pKa = 3.68LVEE194 pKa = 4.46TVAEE198 pKa = 4.25SLEE201 pKa = 3.93HH202 pKa = 7.02LIDD205 pKa = 4.02GAGSHH210 pKa = 5.73QVAQASTLSGVYY222 pKa = 9.59VNGVAVEE229 pKa = 3.99QAPSLEE235 pKa = 5.0DD236 pKa = 3.13YY237 pKa = 11.09HH238 pKa = 9.11SFDD241 pKa = 4.16EE242 pKa = 5.43GEE244 pKa = 4.29DD245 pKa = 3.3DD246 pKa = 4.11GAIVEE251 pKa = 4.64AEE253 pKa = 3.76WGGEE257 pKa = 4.14GSQPPPASVTITMGDD272 pKa = 3.18NTLVNDD278 pKa = 4.03VLVKK282 pKa = 10.73NLWTAAKK289 pKa = 8.48VTVVQGDD296 pKa = 4.04HH297 pKa = 6.68VEE299 pKa = 4.31VNAVVQINAVWDD311 pKa = 3.71TDD313 pKa = 4.51SIGSTVAGWGASNSANDD330 pKa = 4.17LFNIASFDD338 pKa = 3.84HH339 pKa = 7.49DD340 pKa = 4.78DD341 pKa = 3.93SAGSQQLAGASRR353 pKa = 11.84PDD355 pKa = 4.3LYY357 pKa = 10.51PSYY360 pKa = 10.74WNVTTVTGDD369 pKa = 3.35LMIVNWIEE377 pKa = 3.59QLIFMSDD384 pKa = 2.98ADD386 pKa = 3.93MGIVSASGAYY396 pKa = 8.37STIISGDD403 pKa = 3.27NTAVNHH409 pKa = 4.6TTITEE414 pKa = 4.14LGFGYY419 pKa = 10.59DD420 pKa = 4.11LIIIGGSVYY429 pKa = 10.48DD430 pKa = 4.16ANIIQQVNVLFDD442 pKa = 3.74NDD444 pKa = 3.97AVTTTDD450 pKa = 3.05GFEE453 pKa = 4.03SAEE456 pKa = 4.17AGTVSASGNLLWNQASIYY474 pKa = 9.68TIGAAEE480 pKa = 4.13RR481 pKa = 11.84FGEE484 pKa = 4.46LSDD487 pKa = 3.97AYY489 pKa = 10.28RR490 pKa = 11.84DD491 pKa = 3.75AVASLQSGSANLSDD505 pKa = 4.57EE506 pKa = 4.3VLHH509 pKa = 7.12DD510 pKa = 3.75SAFAGLAGLRR520 pKa = 11.84VLHH523 pKa = 5.72VQGDD527 pKa = 4.74FINVQYY533 pKa = 10.74IKK535 pKa = 9.15QTSIIGDD542 pKa = 3.4NDD544 pKa = 3.8QILLAMDD551 pKa = 5.22ALTPNNDD558 pKa = 3.52ASWTVQAGGNTLINNAAILDD578 pKa = 4.0LDD580 pKa = 4.34SFGKK584 pKa = 9.18TYY586 pKa = 11.01VGGEE590 pKa = 4.09QYY592 pKa = 10.88SQEE595 pKa = 4.17TLIQANFISSQPEE608 pKa = 4.12LASHH612 pKa = 7.68DD613 pKa = 4.3PGTLATEE620 pKa = 4.18AVLFLDD626 pKa = 4.21DD627 pKa = 4.34TMLEE631 pKa = 4.16ADD633 pKa = 4.37PAPEE637 pKa = 4.81GGSYY641 pKa = 9.21WPADD645 pKa = 3.59SAGGQDD651 pKa = 5.78DD652 pKa = 4.28GLQTLLAA659 pKa = 4.9
Molecular weight: 69.61 kDa
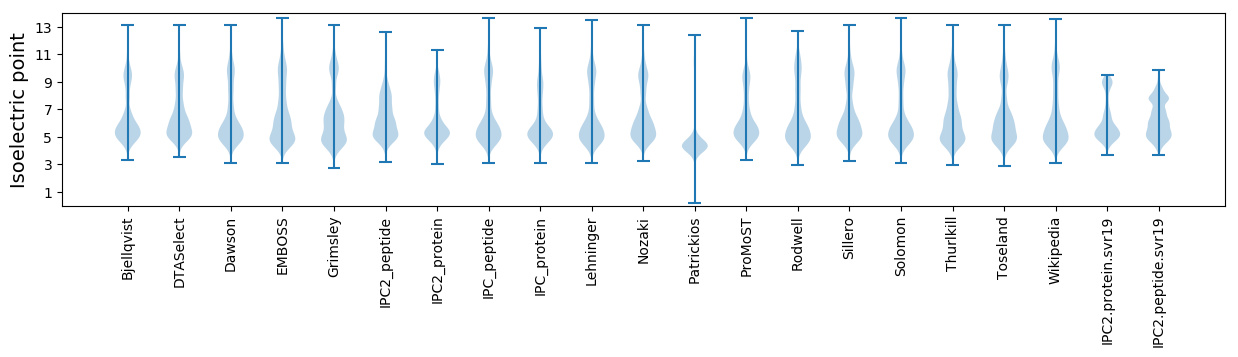
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A081M8A6|A0A081M8A6_9RHIZ Chemotaxis protein CheY OS=Pseudorhizobium pelagicum OX=1509405 GN=GV67_19610 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 10.06LLSPMQSLPVRR13 pKa = 11.84AQQISPPLSRR23 pKa = 11.84PPSPGTRR30 pKa = 11.84PQPPLSQPLLPPSRR44 pKa = 11.84RR45 pKa = 11.84PLPLRR50 pKa = 11.84WPQPPRR56 pKa = 11.84PPPRR60 pKa = 11.84QPQSRR65 pKa = 11.84LRR67 pKa = 11.84KK68 pKa = 9.03PFPLQPPQPLLKK80 pKa = 10.01LQRR83 pKa = 11.84PFPPQLQPLIPMRR96 pKa = 11.84LPRR99 pKa = 11.84PRR101 pKa = 11.84PP102 pKa = 3.36
MM1 pKa = 7.55KK2 pKa = 10.06LLSPMQSLPVRR13 pKa = 11.84AQQISPPLSRR23 pKa = 11.84PPSPGTRR30 pKa = 11.84PQPPLSQPLLPPSRR44 pKa = 11.84RR45 pKa = 11.84PLPLRR50 pKa = 11.84WPQPPRR56 pKa = 11.84PPPRR60 pKa = 11.84QPQSRR65 pKa = 11.84LRR67 pKa = 11.84KK68 pKa = 9.03PFPLQPPQPLLKK80 pKa = 10.01LQRR83 pKa = 11.84PFPPQLQPLIPMRR96 pKa = 11.84LPRR99 pKa = 11.84PRR101 pKa = 11.84PP102 pKa = 3.36
Molecular weight: 11.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1347192 |
37 |
2827 |
305.0 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.904 ± 0.049 | 0.79 ± 0.011 |
5.763 ± 0.031 | 5.962 ± 0.033 |
3.848 ± 0.025 | 8.393 ± 0.038 |
2.016 ± 0.02 | 5.53 ± 0.024 |
3.302 ± 0.03 | 10.094 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.694 ± 0.018 | 2.64 ± 0.018 |
4.95 ± 0.025 | 3.241 ± 0.021 |
6.776 ± 0.043 | 5.705 ± 0.03 |
5.374 ± 0.023 | 7.462 ± 0.025 |
1.266 ± 0.015 | 2.281 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
