
Lachnospiraceae bacterium 3-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
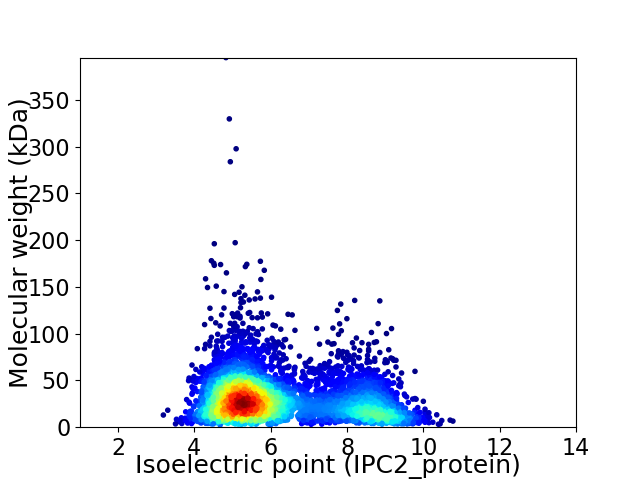
Virtual 2D-PAGE plot for 4984 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
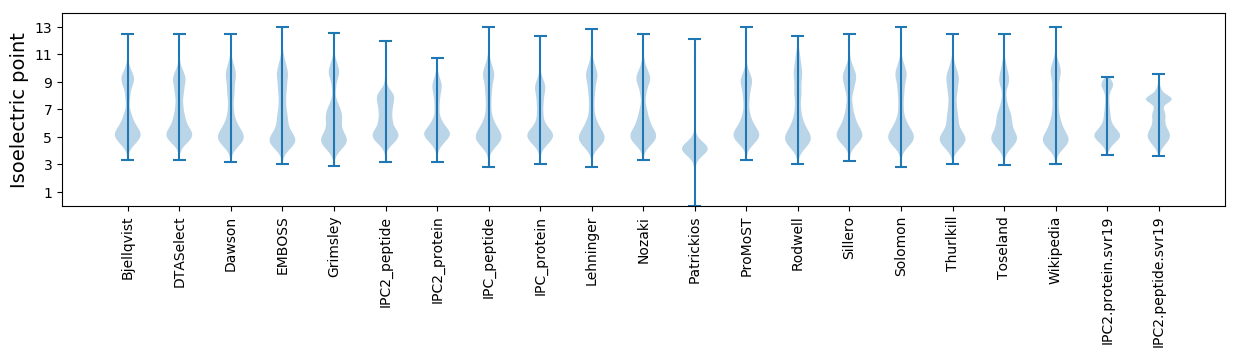
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9IUF5|R9IUF5_9FIRM 23S rRNA (-C5)-methyltransferase OS=Lachnospiraceae bacterium 3-1 OX=397288 GN=C806_01682 PE=4 SV=1
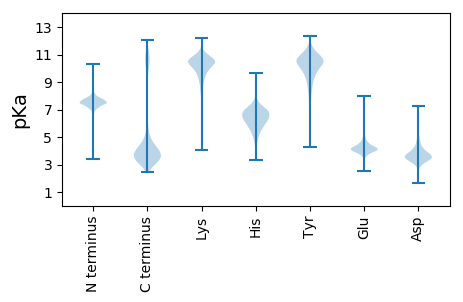
MM1 pKa = 7.18KK2 pKa = 10.27RR3 pKa = 11.84KK4 pKa = 9.27IVSVLMASVMAAALLAGCGGNSNAPSNSDD33 pKa = 3.21SAGNEE38 pKa = 3.86EE39 pKa = 4.4KK40 pKa = 10.76SAGEE44 pKa = 4.01KK45 pKa = 10.28ADD47 pKa = 3.66SKK49 pKa = 11.28GGEE52 pKa = 4.34TVSGDD57 pKa = 3.43GQTLTIVYY65 pKa = 10.24NDD67 pKa = 3.86SNDD70 pKa = 3.57NGDD73 pKa = 3.73NSPVYY78 pKa = 10.04QWINQTYY85 pKa = 8.88EE86 pKa = 3.45NWEE89 pKa = 4.04KK90 pKa = 10.98KK91 pKa = 10.61DD92 pKa = 3.69EE93 pKa = 4.3VKK95 pKa = 10.94LDD97 pKa = 3.43IQALVATDD105 pKa = 3.26SDD107 pKa = 5.12YY108 pKa = 10.67VTKK111 pKa = 10.49IQTLVQDD118 pKa = 5.32DD119 pKa = 4.45STCPDD124 pKa = 4.15LFLEE128 pKa = 5.41DD129 pKa = 3.74TFQLNTDD136 pKa = 3.35VAAGYY141 pKa = 9.99VADD144 pKa = 4.39ITDD147 pKa = 4.51KK148 pKa = 10.6VAGWEE153 pKa = 4.13DD154 pKa = 3.52WNSAIIEE161 pKa = 4.17ALKK164 pKa = 10.51QATSGTDD171 pKa = 3.03GNTYY175 pKa = 10.56AVPISTDD182 pKa = 3.69VRR184 pKa = 11.84GLWYY188 pKa = 10.9NKK190 pKa = 10.27DD191 pKa = 3.32VFEE194 pKa = 4.38AAGLGRR200 pKa = 11.84EE201 pKa = 4.21WQPEE205 pKa = 3.91SWQDD209 pKa = 3.76ILDD212 pKa = 3.69ACAAIKK218 pKa = 9.06EE219 pKa = 4.26KK220 pKa = 11.29CEE222 pKa = 4.29DD223 pKa = 3.77DD224 pKa = 4.45VVPIWFACNSGEE236 pKa = 5.04AEE238 pKa = 4.06ATSMNTFQMLLAGTGDD254 pKa = 4.11SLCDD258 pKa = 3.84DD259 pKa = 3.48ATGVYY264 pKa = 9.9NLSSDD269 pKa = 3.78GVKK272 pKa = 10.66DD273 pKa = 3.28SLGFIKK279 pKa = 9.82TCLDD283 pKa = 3.16EE284 pKa = 6.2GYY286 pKa = 10.41LGTLSEE292 pKa = 4.7ILDD295 pKa = 4.1PSDD298 pKa = 3.13YY299 pKa = 10.72TYY301 pKa = 11.59ANQYY305 pKa = 9.41MSTAKK310 pKa = 10.6LGMYY314 pKa = 10.72LNGSWGYY321 pKa = 11.3NDD323 pKa = 4.04YY324 pKa = 11.0LQDD327 pKa = 3.52SGYY330 pKa = 10.04PMQGYY335 pKa = 7.85TDD337 pKa = 3.81EE338 pKa = 4.52TLSDD342 pKa = 3.71ALGFAQMPTQEE353 pKa = 4.01KK354 pKa = 9.61GGYY357 pKa = 7.25VTMSGGWSWAIANNSDD373 pKa = 3.69QKK375 pKa = 11.4DD376 pKa = 3.56LSFEE380 pKa = 4.65FITEE384 pKa = 4.13LMKK387 pKa = 10.48SDD389 pKa = 3.91NYY391 pKa = 8.78ITYY394 pKa = 9.97IEE396 pKa = 4.43GTGNLATRR404 pKa = 11.84NDD406 pKa = 3.63MMNFDD411 pKa = 4.85AYY413 pKa = 10.54SSRR416 pKa = 11.84MYY418 pKa = 10.32IDD420 pKa = 4.0EE421 pKa = 4.3ATAMSEE427 pKa = 3.94NAFFRR432 pKa = 11.84PHH434 pKa = 7.14DD435 pKa = 3.87EE436 pKa = 4.35NYY438 pKa = 10.91SKK440 pKa = 11.02VSSYY444 pKa = 10.75LYY446 pKa = 10.87EE447 pKa = 4.11MVDD450 pKa = 3.23TVIRR454 pKa = 11.84NNTDD458 pKa = 2.95TEE460 pKa = 4.41TALSDD465 pKa = 3.99FKK467 pKa = 11.16KK468 pKa = 10.78GAATAIGEE476 pKa = 4.21EE477 pKa = 4.67LIQQ480 pKa = 4.7
MM1 pKa = 7.18KK2 pKa = 10.27RR3 pKa = 11.84KK4 pKa = 9.27IVSVLMASVMAAALLAGCGGNSNAPSNSDD33 pKa = 3.21SAGNEE38 pKa = 3.86EE39 pKa = 4.4KK40 pKa = 10.76SAGEE44 pKa = 4.01KK45 pKa = 10.28ADD47 pKa = 3.66SKK49 pKa = 11.28GGEE52 pKa = 4.34TVSGDD57 pKa = 3.43GQTLTIVYY65 pKa = 10.24NDD67 pKa = 3.86SNDD70 pKa = 3.57NGDD73 pKa = 3.73NSPVYY78 pKa = 10.04QWINQTYY85 pKa = 8.88EE86 pKa = 3.45NWEE89 pKa = 4.04KK90 pKa = 10.98KK91 pKa = 10.61DD92 pKa = 3.69EE93 pKa = 4.3VKK95 pKa = 10.94LDD97 pKa = 3.43IQALVATDD105 pKa = 3.26SDD107 pKa = 5.12YY108 pKa = 10.67VTKK111 pKa = 10.49IQTLVQDD118 pKa = 5.32DD119 pKa = 4.45STCPDD124 pKa = 4.15LFLEE128 pKa = 5.41DD129 pKa = 3.74TFQLNTDD136 pKa = 3.35VAAGYY141 pKa = 9.99VADD144 pKa = 4.39ITDD147 pKa = 4.51KK148 pKa = 10.6VAGWEE153 pKa = 4.13DD154 pKa = 3.52WNSAIIEE161 pKa = 4.17ALKK164 pKa = 10.51QATSGTDD171 pKa = 3.03GNTYY175 pKa = 10.56AVPISTDD182 pKa = 3.69VRR184 pKa = 11.84GLWYY188 pKa = 10.9NKK190 pKa = 10.27DD191 pKa = 3.32VFEE194 pKa = 4.38AAGLGRR200 pKa = 11.84EE201 pKa = 4.21WQPEE205 pKa = 3.91SWQDD209 pKa = 3.76ILDD212 pKa = 3.69ACAAIKK218 pKa = 9.06EE219 pKa = 4.26KK220 pKa = 11.29CEE222 pKa = 4.29DD223 pKa = 3.77DD224 pKa = 4.45VVPIWFACNSGEE236 pKa = 5.04AEE238 pKa = 4.06ATSMNTFQMLLAGTGDD254 pKa = 4.11SLCDD258 pKa = 3.84DD259 pKa = 3.48ATGVYY264 pKa = 9.9NLSSDD269 pKa = 3.78GVKK272 pKa = 10.66DD273 pKa = 3.28SLGFIKK279 pKa = 9.82TCLDD283 pKa = 3.16EE284 pKa = 6.2GYY286 pKa = 10.41LGTLSEE292 pKa = 4.7ILDD295 pKa = 4.1PSDD298 pKa = 3.13YY299 pKa = 10.72TYY301 pKa = 11.59ANQYY305 pKa = 9.41MSTAKK310 pKa = 10.6LGMYY314 pKa = 10.72LNGSWGYY321 pKa = 11.3NDD323 pKa = 4.04YY324 pKa = 11.0LQDD327 pKa = 3.52SGYY330 pKa = 10.04PMQGYY335 pKa = 7.85TDD337 pKa = 3.81EE338 pKa = 4.52TLSDD342 pKa = 3.71ALGFAQMPTQEE353 pKa = 4.01KK354 pKa = 9.61GGYY357 pKa = 7.25VTMSGGWSWAIANNSDD373 pKa = 3.69QKK375 pKa = 11.4DD376 pKa = 3.56LSFEE380 pKa = 4.65FITEE384 pKa = 4.13LMKK387 pKa = 10.48SDD389 pKa = 3.91NYY391 pKa = 8.78ITYY394 pKa = 9.97IEE396 pKa = 4.43GTGNLATRR404 pKa = 11.84NDD406 pKa = 3.63MMNFDD411 pKa = 4.85AYY413 pKa = 10.54SSRR416 pKa = 11.84MYY418 pKa = 10.32IDD420 pKa = 4.0EE421 pKa = 4.3ATAMSEE427 pKa = 3.94NAFFRR432 pKa = 11.84PHH434 pKa = 7.14DD435 pKa = 3.87EE436 pKa = 4.35NYY438 pKa = 10.91SKK440 pKa = 11.02VSSYY444 pKa = 10.75LYY446 pKa = 10.87EE447 pKa = 4.11MVDD450 pKa = 3.23TVIRR454 pKa = 11.84NNTDD458 pKa = 2.95TEE460 pKa = 4.41TALSDD465 pKa = 3.99FKK467 pKa = 11.16KK468 pKa = 10.78GAATAIGEE476 pKa = 4.21EE477 pKa = 4.67LIQQ480 pKa = 4.7
Molecular weight: 52.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9INK9|R9INK9_9FIRM Excisionase family DNA binding domain-containing protein OS=Lachnospiraceae bacterium 3-1 OX=397288 GN=C806_03001 PE=4 SV=1
MM1 pKa = 7.84AGMVLRR7 pKa = 11.84KK8 pKa = 8.44TGVVLLTILKK18 pKa = 9.45WILQAALVALKK29 pKa = 10.49LVLGIAKK36 pKa = 10.15LFLLLLSLVMRR47 pKa = 11.84IVLTMIGVSVGRR59 pKa = 11.84RR60 pKa = 3.23
MM1 pKa = 7.84AGMVLRR7 pKa = 11.84KK8 pKa = 8.44TGVVLLTILKK18 pKa = 9.45WILQAALVALKK29 pKa = 10.49LVLGIAKK36 pKa = 10.15LFLLLLSLVMRR47 pKa = 11.84IVLTMIGVSVGRR59 pKa = 11.84RR60 pKa = 3.23
Molecular weight: 6.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1447276 |
20 |
3497 |
290.4 |
32.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.08 ± 0.038 | 1.506 ± 0.012 |
5.28 ± 0.029 | 8.256 ± 0.041 |
4.125 ± 0.025 | 6.923 ± 0.036 |
1.784 ± 0.017 | 7.249 ± 0.034 |
7.303 ± 0.033 | 8.815 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.133 ± 0.018 | 4.475 ± 0.024 |
3.158 ± 0.024 | 3.725 ± 0.026 |
4.529 ± 0.028 | 5.849 ± 0.027 |
5.165 ± 0.029 | 6.471 ± 0.031 |
0.966 ± 0.013 | 4.207 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
