
Mizuhopecten yessoensis (Japanese scallop) (Patinopecten yessoensis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Bivalvia; Autobranchia; Pteriomorphia; Pectinida; Pectinoidea; Pectinidae; Mizuhopecten
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
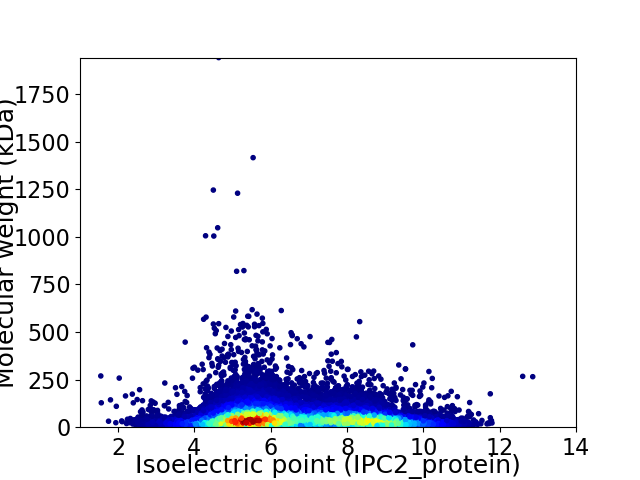
Virtual 2D-PAGE plot for 22409 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A210PL05|A0A210PL05_MIZYE Pleckstrin homology domain-containing family G member 5 OS=Mizuhopecten yessoensis OX=6573 GN=KP79_PYT21310 PE=4 SV=1
MM1 pKa = 7.5LNVCKK6 pKa = 10.36IIPPFSRR13 pKa = 11.84SVQSWVGDD21 pKa = 3.67VICIGTLLIVRR32 pKa = 11.84RR33 pKa = 11.84QSRR36 pKa = 11.84DD37 pKa = 2.55GDD39 pKa = 4.12VICIGILLIVRR50 pKa = 11.84RR51 pKa = 11.84QSWDD55 pKa = 2.99GDD57 pKa = 4.13VICIDD62 pKa = 4.12GDD64 pKa = 4.02IICIDD69 pKa = 3.81GDD71 pKa = 4.44VICLGILLIVRR82 pKa = 11.84RR83 pKa = 11.84QSWDD87 pKa = 3.24GNVICIDD94 pKa = 3.76GDD96 pKa = 4.1VICIDD101 pKa = 4.12GDD103 pKa = 4.15VICIDD108 pKa = 3.61VDD110 pKa = 4.17VICIDD115 pKa = 4.15GDD117 pKa = 3.89INCIDD122 pKa = 4.32GDD124 pKa = 4.4VICIDD129 pKa = 3.83GDD131 pKa = 3.99IICILIFLIVRR142 pKa = 11.84RR143 pKa = 11.84QSMDD147 pKa = 2.86GDD149 pKa = 4.34VICIDD154 pKa = 4.21GDD156 pKa = 3.98VLCIDD161 pKa = 4.62GDD163 pKa = 4.29VICIDD168 pKa = 4.12GDD170 pKa = 4.15VICIDD175 pKa = 3.61VDD177 pKa = 4.17VICIDD182 pKa = 4.88GDD184 pKa = 4.22IIWIDD189 pKa = 3.43GDD191 pKa = 4.44VICLGILLIVRR202 pKa = 11.84RR203 pKa = 11.84QSWAGDD209 pKa = 3.69VICIDD214 pKa = 4.1GDD216 pKa = 4.0IICIGIFLIVRR227 pKa = 11.84RR228 pKa = 11.84QSRR231 pKa = 11.84DD232 pKa = 2.62GDD234 pKa = 4.15VICIDD239 pKa = 4.12GDD241 pKa = 4.11VICIDD246 pKa = 3.83GDD248 pKa = 3.99IICILIFLIVRR259 pKa = 11.84RR260 pKa = 11.84QSMDD264 pKa = 2.86GDD266 pKa = 4.34VICIDD271 pKa = 3.95GDD273 pKa = 4.07VICIRR278 pKa = 11.84IFLIVRR284 pKa = 11.84RR285 pKa = 11.84QSWDD289 pKa = 2.8GDD291 pKa = 3.82VIYY294 pKa = 10.39IGILLIVRR302 pKa = 11.84RR303 pKa = 11.84QSWDD307 pKa = 2.89GDD309 pKa = 4.1VICIRR314 pKa = 11.84ILLIVRR320 pKa = 11.84RR321 pKa = 11.84QSWDD325 pKa = 2.86GDD327 pKa = 4.11VII329 pKa = 4.0
MM1 pKa = 7.5LNVCKK6 pKa = 10.36IIPPFSRR13 pKa = 11.84SVQSWVGDD21 pKa = 3.67VICIGTLLIVRR32 pKa = 11.84RR33 pKa = 11.84QSRR36 pKa = 11.84DD37 pKa = 2.55GDD39 pKa = 4.12VICIGILLIVRR50 pKa = 11.84RR51 pKa = 11.84QSWDD55 pKa = 2.99GDD57 pKa = 4.13VICIDD62 pKa = 4.12GDD64 pKa = 4.02IICIDD69 pKa = 3.81GDD71 pKa = 4.44VICLGILLIVRR82 pKa = 11.84RR83 pKa = 11.84QSWDD87 pKa = 3.24GNVICIDD94 pKa = 3.76GDD96 pKa = 4.1VICIDD101 pKa = 4.12GDD103 pKa = 4.15VICIDD108 pKa = 3.61VDD110 pKa = 4.17VICIDD115 pKa = 4.15GDD117 pKa = 3.89INCIDD122 pKa = 4.32GDD124 pKa = 4.4VICIDD129 pKa = 3.83GDD131 pKa = 3.99IICILIFLIVRR142 pKa = 11.84RR143 pKa = 11.84QSMDD147 pKa = 2.86GDD149 pKa = 4.34VICIDD154 pKa = 4.21GDD156 pKa = 3.98VLCIDD161 pKa = 4.62GDD163 pKa = 4.29VICIDD168 pKa = 4.12GDD170 pKa = 4.15VICIDD175 pKa = 3.61VDD177 pKa = 4.17VICIDD182 pKa = 4.88GDD184 pKa = 4.22IIWIDD189 pKa = 3.43GDD191 pKa = 4.44VICLGILLIVRR202 pKa = 11.84RR203 pKa = 11.84QSWAGDD209 pKa = 3.69VICIDD214 pKa = 4.1GDD216 pKa = 4.0IICIGIFLIVRR227 pKa = 11.84RR228 pKa = 11.84QSRR231 pKa = 11.84DD232 pKa = 2.62GDD234 pKa = 4.15VICIDD239 pKa = 4.12GDD241 pKa = 4.11VICIDD246 pKa = 3.83GDD248 pKa = 3.99IICILIFLIVRR259 pKa = 11.84RR260 pKa = 11.84QSMDD264 pKa = 2.86GDD266 pKa = 4.34VICIDD271 pKa = 3.95GDD273 pKa = 4.07VICIRR278 pKa = 11.84IFLIVRR284 pKa = 11.84RR285 pKa = 11.84QSWDD289 pKa = 2.8GDD291 pKa = 3.82VIYY294 pKa = 10.39IGILLIVRR302 pKa = 11.84RR303 pKa = 11.84QSWDD307 pKa = 2.89GDD309 pKa = 4.1VICIRR314 pKa = 11.84ILLIVRR320 pKa = 11.84RR321 pKa = 11.84QSWDD325 pKa = 2.86GDD327 pKa = 4.11VII329 pKa = 4.0
Molecular weight: 36.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A210PP45|A0A210PP45_MIZYE Multidrug resistance-associated protein 7 OS=Mizuhopecten yessoensis OX=6573 GN=KP79_PYT10635 PE=4 SV=1
MM1 pKa = 7.27RR2 pKa = 11.84TIGTMTMRR10 pKa = 11.84TIGTMSVRR18 pKa = 11.84TIGTMTMRR26 pKa = 11.84TIGTMTMRR34 pKa = 11.84TIGTMSVRR42 pKa = 11.84TIGTMTMRR50 pKa = 11.84TIGTMTMRR58 pKa = 11.84TIGTMSVRR66 pKa = 11.84TIGTMTMRR74 pKa = 11.84TIGTMTMRR82 pKa = 11.84TIGKK86 pKa = 8.59MSVRR90 pKa = 11.84TIGTMTLRR98 pKa = 11.84KK99 pKa = 9.37IGQQ102 pKa = 3.19
MM1 pKa = 7.27RR2 pKa = 11.84TIGTMTMRR10 pKa = 11.84TIGTMSVRR18 pKa = 11.84TIGTMTMRR26 pKa = 11.84TIGTMTMRR34 pKa = 11.84TIGTMSVRR42 pKa = 11.84TIGTMTMRR50 pKa = 11.84TIGTMTMRR58 pKa = 11.84TIGTMSVRR66 pKa = 11.84TIGTMTMRR74 pKa = 11.84TIGTMTMRR82 pKa = 11.84TIGKK86 pKa = 8.59MSVRR90 pKa = 11.84TIGTMTLRR98 pKa = 11.84KK99 pKa = 9.37IGQQ102 pKa = 3.19
Molecular weight: 11.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11322112 |
45 |
18501 |
505.2 |
56.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.426 ± 0.015 | 2.263 ± 0.021 |
5.907 ± 0.018 | 5.986 ± 0.024 |
3.702 ± 0.015 | 5.986 ± 0.034 |
2.611 ± 0.014 | 5.423 ± 0.018 |
6.105 ± 0.023 | 8.483 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.717 ± 0.011 | 4.828 ± 0.017 |
4.907 ± 0.022 | 4.266 ± 0.018 |
5.095 ± 0.021 | 8.364 ± 0.029 |
6.851 ± 0.029 | 6.683 ± 0.023 |
1.069 ± 0.008 | 3.325 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
