
Fowl aviadenovirus 6
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus; Fowl aviadenovirus E
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
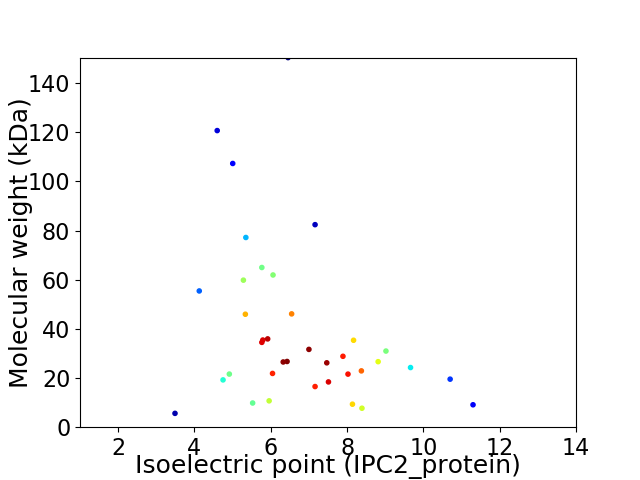
Virtual 2D-PAGE plot for 37 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
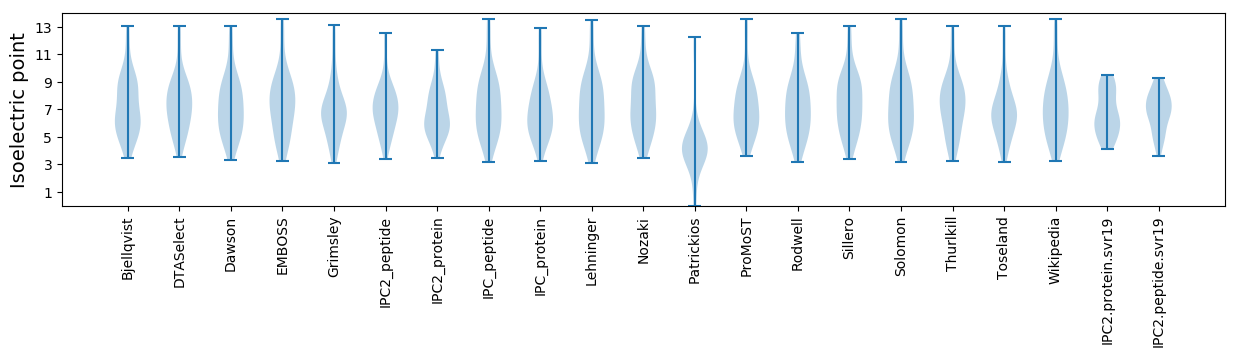
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191ULR9|A0A191ULR9_9ADEN Penton protein OS=Fowl aviadenovirus 6 OX=172862 GN=L2 PE=2 SV=1
MM1 pKa = 7.12ATSTPHH7 pKa = 6.67AFSFGQIGSRR17 pKa = 11.84KK18 pKa = 9.34RR19 pKa = 11.84PAGGDD24 pKa = 3.67GEE26 pKa = 4.79RR27 pKa = 11.84DD28 pKa = 3.22ASKK31 pKa = 10.42VPKK34 pKa = 8.97MQTPAPSATANGNDD48 pKa = 4.13EE49 pKa = 5.04LDD51 pKa = 3.4LVYY54 pKa = 10.27PFWLQNGSTGGGGGGGSGGNPSLNPPFLDD83 pKa = 3.61PNGPLAVQNNLLKK96 pKa = 11.07VNTAAPITVANKK108 pKa = 10.48AITLAYY114 pKa = 10.26DD115 pKa = 3.61PEE117 pKa = 4.37SLEE120 pKa = 5.58LNNQQQLSVKK130 pKa = 9.59IDD132 pKa = 3.39PEE134 pKa = 5.14GPLKK138 pKa = 9.55PTADD142 pKa = 5.01GIQLSVDD149 pKa = 3.44PTTLEE154 pKa = 3.5VDD156 pKa = 3.24ADD158 pKa = 3.8DD159 pKa = 4.76WEE161 pKa = 4.57LTVNLDD167 pKa = 4.43LNGPLDD173 pKa = 3.78ATASGITVRR182 pKa = 11.84VDD184 pKa = 3.02DD185 pKa = 4.39TLLIEE190 pKa = 6.01DD191 pKa = 5.13DD192 pKa = 5.2DD193 pKa = 4.7SGQGKK198 pKa = 8.91EE199 pKa = 4.5LGVNLNPAGPITADD213 pKa = 3.33QNGLDD218 pKa = 5.32LEE220 pKa = 5.95IDD222 pKa = 3.82DD223 pKa = 4.21QTLKK227 pKa = 11.0VNSGTSGGVLAVQIKK242 pKa = 8.81PQGGLTSQTDD252 pKa = 5.22GIQVSTQNSITLNNGALDD270 pKa = 3.92VKK272 pKa = 10.74VVADD276 pKa = 4.45GPLASTANGLTLNYY290 pKa = 9.83DD291 pKa = 3.5PSDD294 pKa = 3.54FTVNDD299 pKa = 3.85AGTLSILRR307 pKa = 11.84NPSVVANAYY316 pKa = 7.22LTSGASTLQQFTAQGEE332 pKa = 4.3NSSQFSFPCAYY343 pKa = 10.45YY344 pKa = 9.99LQQWLSDD351 pKa = 3.44GLIFSSLYY359 pKa = 10.6LKK361 pKa = 10.49LDD363 pKa = 3.79RR364 pKa = 11.84AHH366 pKa = 6.13FTHH369 pKa = 6.32MQTGDD374 pKa = 2.99TYY376 pKa = 11.7QNAKK380 pKa = 10.5YY381 pKa = 8.31FTFWVGAGTSFNLSTLTVPTITPNTTQWNAFEE413 pKa = 4.54PALDD417 pKa = 3.89YY418 pKa = 11.51SGAPPFDD425 pKa = 3.92YY426 pKa = 10.94LSTSVLTIYY435 pKa = 10.58FEE437 pKa = 4.28PTTGRR442 pKa = 11.84LEE444 pKa = 4.18SYY446 pKa = 10.9LPVLTGNWSQTPYY459 pKa = 11.18NPGTITLCVRR469 pKa = 11.84AVRR472 pKa = 11.84VQSRR476 pKa = 11.84SQQTFSTLVCYY487 pKa = 10.25NFRR490 pKa = 11.84CQNAGIFNSNATAGTMTLGPIFFSCPAVSTVNAPP524 pKa = 3.17
MM1 pKa = 7.12ATSTPHH7 pKa = 6.67AFSFGQIGSRR17 pKa = 11.84KK18 pKa = 9.34RR19 pKa = 11.84PAGGDD24 pKa = 3.67GEE26 pKa = 4.79RR27 pKa = 11.84DD28 pKa = 3.22ASKK31 pKa = 10.42VPKK34 pKa = 8.97MQTPAPSATANGNDD48 pKa = 4.13EE49 pKa = 5.04LDD51 pKa = 3.4LVYY54 pKa = 10.27PFWLQNGSTGGGGGGGSGGNPSLNPPFLDD83 pKa = 3.61PNGPLAVQNNLLKK96 pKa = 11.07VNTAAPITVANKK108 pKa = 10.48AITLAYY114 pKa = 10.26DD115 pKa = 3.61PEE117 pKa = 4.37SLEE120 pKa = 5.58LNNQQQLSVKK130 pKa = 9.59IDD132 pKa = 3.39PEE134 pKa = 5.14GPLKK138 pKa = 9.55PTADD142 pKa = 5.01GIQLSVDD149 pKa = 3.44PTTLEE154 pKa = 3.5VDD156 pKa = 3.24ADD158 pKa = 3.8DD159 pKa = 4.76WEE161 pKa = 4.57LTVNLDD167 pKa = 4.43LNGPLDD173 pKa = 3.78ATASGITVRR182 pKa = 11.84VDD184 pKa = 3.02DD185 pKa = 4.39TLLIEE190 pKa = 6.01DD191 pKa = 5.13DD192 pKa = 5.2DD193 pKa = 4.7SGQGKK198 pKa = 8.91EE199 pKa = 4.5LGVNLNPAGPITADD213 pKa = 3.33QNGLDD218 pKa = 5.32LEE220 pKa = 5.95IDD222 pKa = 3.82DD223 pKa = 4.21QTLKK227 pKa = 11.0VNSGTSGGVLAVQIKK242 pKa = 8.81PQGGLTSQTDD252 pKa = 5.22GIQVSTQNSITLNNGALDD270 pKa = 3.92VKK272 pKa = 10.74VVADD276 pKa = 4.45GPLASTANGLTLNYY290 pKa = 9.83DD291 pKa = 3.5PSDD294 pKa = 3.54FTVNDD299 pKa = 3.85AGTLSILRR307 pKa = 11.84NPSVVANAYY316 pKa = 7.22LTSGASTLQQFTAQGEE332 pKa = 4.3NSSQFSFPCAYY343 pKa = 10.45YY344 pKa = 9.99LQQWLSDD351 pKa = 3.44GLIFSSLYY359 pKa = 10.6LKK361 pKa = 10.49LDD363 pKa = 3.79RR364 pKa = 11.84AHH366 pKa = 6.13FTHH369 pKa = 6.32MQTGDD374 pKa = 2.99TYY376 pKa = 11.7QNAKK380 pKa = 10.5YY381 pKa = 8.31FTFWVGAGTSFNLSTLTVPTITPNTTQWNAFEE413 pKa = 4.54PALDD417 pKa = 3.89YY418 pKa = 11.51SGAPPFDD425 pKa = 3.92YY426 pKa = 10.94LSTSVLTIYY435 pKa = 10.58FEE437 pKa = 4.28PTTGRR442 pKa = 11.84LEE444 pKa = 4.18SYY446 pKa = 10.9LPVLTGNWSQTPYY459 pKa = 11.18NPGTITLCVRR469 pKa = 11.84AVRR472 pKa = 11.84VQSRR476 pKa = 11.84SQQTFSTLVCYY487 pKa = 10.25NFRR490 pKa = 11.84CQNAGIFNSNATAGTMTLGPIFFSCPAVSTVNAPP524 pKa = 3.17
Molecular weight: 55.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191ULS6|A0A191ULS6_9ADEN ORF17 OS=Fowl aviadenovirus 6 OX=172862 PE=4 SV=1
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LTLRR36 pKa = 11.84SLLGLGTVSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GGRR52 pKa = 11.84SSRR55 pKa = 11.84RR56 pKa = 11.84SSRR59 pKa = 11.84PASTTTRR66 pKa = 11.84LMVVRR71 pKa = 11.84TSRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 3.32
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LTLRR36 pKa = 11.84SLLGLGTVSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GGRR52 pKa = 11.84SSRR55 pKa = 11.84RR56 pKa = 11.84SSRR59 pKa = 11.84PASTTTRR66 pKa = 11.84LMVVRR71 pKa = 11.84TSRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 3.32
Molecular weight: 9.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12920 |
54 |
1305 |
349.2 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.274 ± 0.437 | 1.827 ± 0.205 |
5.704 ± 0.273 | 6.099 ± 0.442 |
4.149 ± 0.238 | 6.378 ± 0.322 |
2.337 ± 0.192 | 3.576 ± 0.155 |
3.46 ± 0.273 | 8.994 ± 0.344 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.276 ± 0.172 | 3.916 ± 0.401 |
7.036 ± 0.302 | 3.692 ± 0.264 |
7.67 ± 0.453 | 6.966 ± 0.368 |
5.859 ± 0.36 | 6.61 ± 0.289 |
1.37 ± 0.116 | 3.808 ± 0.302 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
