
Capybara microvirus Cap1_SP_162
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
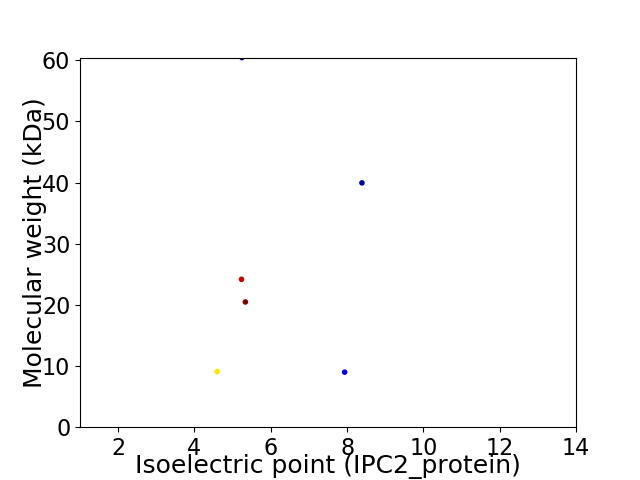
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W816|A0A4P8W816_9VIRU Nonstructural protein OS=Capybara microvirus Cap1_SP_162 OX=2585398 PE=4 SV=1
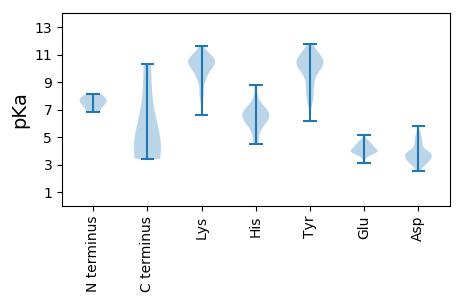
MM1 pKa = 7.62AVYY4 pKa = 10.35KK5 pKa = 10.07IYY7 pKa = 10.76SVQDD11 pKa = 3.14VLIGFNAPFIMINDD25 pKa = 5.17DD26 pKa = 3.11IAKK29 pKa = 9.93RR30 pKa = 11.84EE31 pKa = 4.0YY32 pKa = 11.1RR33 pKa = 11.84NMLEE37 pKa = 4.31TNPNRR42 pKa = 11.84ADD44 pKa = 3.14MRR46 pKa = 11.84LFRR49 pKa = 11.84LASFDD54 pKa = 4.83DD55 pKa = 3.89EE56 pKa = 4.74TGMVTPEE63 pKa = 3.64ITPVLVEE70 pKa = 4.37GGANGSNKK78 pKa = 10.29DD79 pKa = 3.65SVV81 pKa = 3.41
MM1 pKa = 7.62AVYY4 pKa = 10.35KK5 pKa = 10.07IYY7 pKa = 10.76SVQDD11 pKa = 3.14VLIGFNAPFIMINDD25 pKa = 5.17DD26 pKa = 3.11IAKK29 pKa = 9.93RR30 pKa = 11.84EE31 pKa = 4.0YY32 pKa = 11.1RR33 pKa = 11.84NMLEE37 pKa = 4.31TNPNRR42 pKa = 11.84ADD44 pKa = 3.14MRR46 pKa = 11.84LFRR49 pKa = 11.84LASFDD54 pKa = 4.83DD55 pKa = 3.89EE56 pKa = 4.74TGMVTPEE63 pKa = 3.64ITPVLVEE70 pKa = 4.37GGANGSNKK78 pKa = 10.29DD79 pKa = 3.65SVV81 pKa = 3.41
Molecular weight: 9.09 kDa
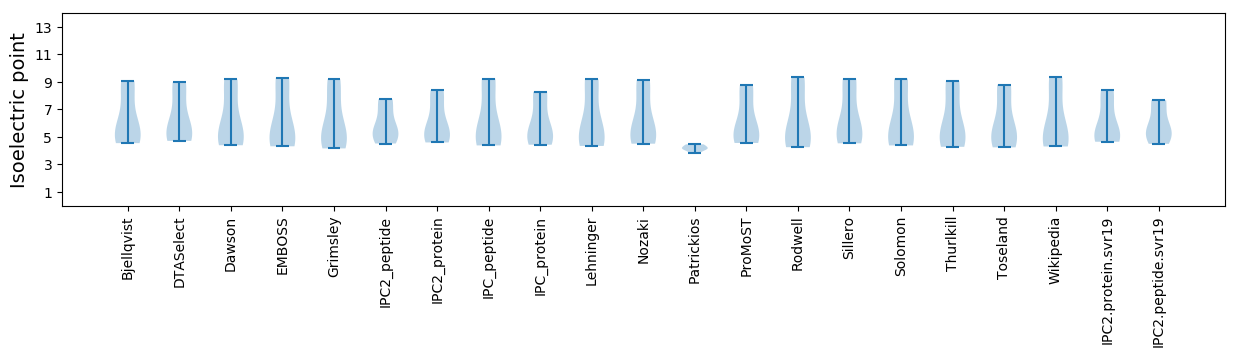
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5B1|A0A4P8W5B1_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_162 OX=2585398 PE=4 SV=1
MM1 pKa = 7.61ARR3 pKa = 11.84LEE5 pKa = 4.86LPGQTYY11 pKa = 10.52INKK14 pKa = 9.45QGKK17 pKa = 9.05KK18 pKa = 6.88SHH20 pKa = 6.27RR21 pKa = 11.84CEE23 pKa = 3.58ITSWADD29 pKa = 3.08VEE31 pKa = 4.41QNPKK35 pKa = 9.99FYY37 pKa = 8.79KK38 pKa = 9.22TKK40 pKa = 8.64YY41 pKa = 9.32HH42 pKa = 6.59RR43 pKa = 11.84VDD45 pKa = 4.88KK46 pKa = 10.55IPCGQCMEE54 pKa = 4.33CRR56 pKa = 11.84LNKK59 pKa = 9.48ARR61 pKa = 11.84QWANRR66 pKa = 11.84IVLEE70 pKa = 4.17KK71 pKa = 10.13QDD73 pKa = 3.83YY74 pKa = 10.46PEE76 pKa = 4.05EE77 pKa = 4.16QVWFLTLTYY86 pKa = 10.57DD87 pKa = 4.56DD88 pKa = 3.88EE89 pKa = 4.66HH90 pKa = 8.8VPFGMTVDD98 pKa = 3.63MEE100 pKa = 4.49TGEE103 pKa = 4.7EE104 pKa = 4.08IVNMTLKK111 pKa = 10.85KK112 pKa = 9.32EE113 pKa = 4.14DD114 pKa = 3.8EE115 pKa = 4.53VKK117 pKa = 10.69FMHH120 pKa = 7.51DD121 pKa = 2.84LRR123 pKa = 11.84QYY125 pKa = 8.02YY126 pKa = 7.61QRR128 pKa = 11.84KK129 pKa = 8.84YY130 pKa = 10.24NIDD133 pKa = 3.75GIRR136 pKa = 11.84FYY138 pKa = 10.73MAGEE142 pKa = 4.16YY143 pKa = 10.88GSTTNRR149 pKa = 11.84PHH151 pKa = 5.34YY152 pKa = 10.46HH153 pKa = 6.18YY154 pKa = 10.43ILYY157 pKa = 10.07GLPLDD162 pKa = 3.6QTKK165 pKa = 10.7LKK167 pKa = 10.28RR168 pKa = 11.84ISINEE173 pKa = 4.02LGQTLWTHH181 pKa = 5.92EE182 pKa = 4.65EE183 pKa = 4.15IEE185 pKa = 4.51KK186 pKa = 10.03IWNKK190 pKa = 10.35GMITIGRR197 pKa = 11.84VTWEE201 pKa = 4.03SACYY205 pKa = 8.4VARR208 pKa = 11.84YY209 pKa = 6.18CTKK212 pKa = 10.74KK213 pKa = 9.16ITGKK217 pKa = 10.29DD218 pKa = 3.29AEE220 pKa = 4.61WYY222 pKa = 9.94YY223 pKa = 10.76KK224 pKa = 10.35AQGIIPPFTQMSRR237 pKa = 11.84RR238 pKa = 11.84PGIGANYY245 pKa = 10.14LEE247 pKa = 4.43KK248 pKa = 11.07NKK250 pKa = 10.57DD251 pKa = 3.47AIYY254 pKa = 8.01TTDD257 pKa = 4.55SIPIANKK264 pKa = 8.51KK265 pKa = 6.57TASLVSPPKK274 pKa = 10.25SFDD277 pKa = 3.85RR278 pKa = 11.84IMEE281 pKa = 4.25KK282 pKa = 10.82YY283 pKa = 9.43EE284 pKa = 3.9PEE286 pKa = 3.82FMEE289 pKa = 3.75QLKK292 pKa = 10.26RR293 pKa = 11.84EE294 pKa = 3.94RR295 pKa = 11.84RR296 pKa = 11.84RR297 pKa = 11.84RR298 pKa = 11.84AEE300 pKa = 3.86LNEE303 pKa = 3.7AAKK306 pKa = 10.59FKK308 pKa = 10.59ATDD311 pKa = 3.59LTPTEE316 pKa = 3.34WRR318 pKa = 11.84YY319 pKa = 10.26QNEE322 pKa = 4.26EE323 pKa = 4.08KK324 pKa = 10.55IKK326 pKa = 10.89ARR328 pKa = 11.84TKK330 pKa = 10.33SLVRR334 pKa = 11.84EE335 pKa = 4.31LL336 pKa = 4.01
MM1 pKa = 7.61ARR3 pKa = 11.84LEE5 pKa = 4.86LPGQTYY11 pKa = 10.52INKK14 pKa = 9.45QGKK17 pKa = 9.05KK18 pKa = 6.88SHH20 pKa = 6.27RR21 pKa = 11.84CEE23 pKa = 3.58ITSWADD29 pKa = 3.08VEE31 pKa = 4.41QNPKK35 pKa = 9.99FYY37 pKa = 8.79KK38 pKa = 9.22TKK40 pKa = 8.64YY41 pKa = 9.32HH42 pKa = 6.59RR43 pKa = 11.84VDD45 pKa = 4.88KK46 pKa = 10.55IPCGQCMEE54 pKa = 4.33CRR56 pKa = 11.84LNKK59 pKa = 9.48ARR61 pKa = 11.84QWANRR66 pKa = 11.84IVLEE70 pKa = 4.17KK71 pKa = 10.13QDD73 pKa = 3.83YY74 pKa = 10.46PEE76 pKa = 4.05EE77 pKa = 4.16QVWFLTLTYY86 pKa = 10.57DD87 pKa = 4.56DD88 pKa = 3.88EE89 pKa = 4.66HH90 pKa = 8.8VPFGMTVDD98 pKa = 3.63MEE100 pKa = 4.49TGEE103 pKa = 4.7EE104 pKa = 4.08IVNMTLKK111 pKa = 10.85KK112 pKa = 9.32EE113 pKa = 4.14DD114 pKa = 3.8EE115 pKa = 4.53VKK117 pKa = 10.69FMHH120 pKa = 7.51DD121 pKa = 2.84LRR123 pKa = 11.84QYY125 pKa = 8.02YY126 pKa = 7.61QRR128 pKa = 11.84KK129 pKa = 8.84YY130 pKa = 10.24NIDD133 pKa = 3.75GIRR136 pKa = 11.84FYY138 pKa = 10.73MAGEE142 pKa = 4.16YY143 pKa = 10.88GSTTNRR149 pKa = 11.84PHH151 pKa = 5.34YY152 pKa = 10.46HH153 pKa = 6.18YY154 pKa = 10.43ILYY157 pKa = 10.07GLPLDD162 pKa = 3.6QTKK165 pKa = 10.7LKK167 pKa = 10.28RR168 pKa = 11.84ISINEE173 pKa = 4.02LGQTLWTHH181 pKa = 5.92EE182 pKa = 4.65EE183 pKa = 4.15IEE185 pKa = 4.51KK186 pKa = 10.03IWNKK190 pKa = 10.35GMITIGRR197 pKa = 11.84VTWEE201 pKa = 4.03SACYY205 pKa = 8.4VARR208 pKa = 11.84YY209 pKa = 6.18CTKK212 pKa = 10.74KK213 pKa = 9.16ITGKK217 pKa = 10.29DD218 pKa = 3.29AEE220 pKa = 4.61WYY222 pKa = 9.94YY223 pKa = 10.76KK224 pKa = 10.35AQGIIPPFTQMSRR237 pKa = 11.84RR238 pKa = 11.84PGIGANYY245 pKa = 10.14LEE247 pKa = 4.43KK248 pKa = 11.07NKK250 pKa = 10.57DD251 pKa = 3.47AIYY254 pKa = 8.01TTDD257 pKa = 4.55SIPIANKK264 pKa = 8.51KK265 pKa = 6.57TASLVSPPKK274 pKa = 10.25SFDD277 pKa = 3.85RR278 pKa = 11.84IMEE281 pKa = 4.25KK282 pKa = 10.82YY283 pKa = 9.43EE284 pKa = 3.9PEE286 pKa = 3.82FMEE289 pKa = 3.75QLKK292 pKa = 10.26RR293 pKa = 11.84EE294 pKa = 3.94RR295 pKa = 11.84RR296 pKa = 11.84RR297 pKa = 11.84RR298 pKa = 11.84AEE300 pKa = 3.86LNEE303 pKa = 3.7AAKK306 pKa = 10.59FKK308 pKa = 10.59ATDD311 pKa = 3.59LTPTEE316 pKa = 3.34WRR318 pKa = 11.84YY319 pKa = 10.26QNEE322 pKa = 4.26EE323 pKa = 4.08KK324 pKa = 10.55IKK326 pKa = 10.89ARR328 pKa = 11.84TKK330 pKa = 10.33SLVRR334 pKa = 11.84EE335 pKa = 4.31LL336 pKa = 4.01
Molecular weight: 39.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1444 |
75 |
536 |
240.7 |
27.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.28 ± 2.379 | 0.9 ± 0.291 |
4.848 ± 0.614 | 8.102 ± 0.992 |
3.116 ± 0.466 | 6.371 ± 0.625 |
1.385 ± 0.339 | 6.648 ± 0.559 |
6.717 ± 1.2 | 6.025 ± 0.421 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.047 ± 0.311 | 4.848 ± 0.45 |
4.155 ± 0.84 | 4.432 ± 0.379 |
5.263 ± 0.834 | 7.271 ± 2.116 |
6.925 ± 0.764 | 3.74 ± 0.562 |
1.87 ± 0.314 | 5.055 ± 0.576 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
