
Pseudodesulfovibrio aespoeensis (strain ATCC 700646 / DSM 10631 / Aspo-2) (Desulfovibrio aespoeensis)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Pseudodesulfovibrio; Pseudodesulfovibrio aespoeensis
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
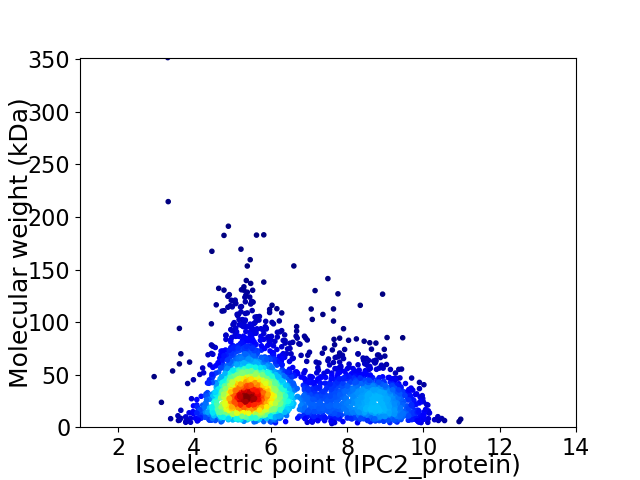
Virtual 2D-PAGE plot for 3269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
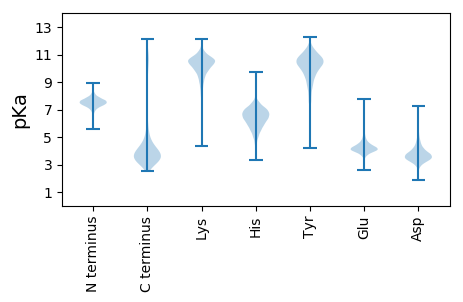
Protein with the lowest isoelectric point:
>tr|E6VTH3|E6VTH3_PSEA9 Sigma-54 factor interaction domain-containing protein OS=Pseudodesulfovibrio aespoeensis (strain ATCC 700646 / DSM 10631 / Aspo-2) OX=643562 GN=Daes_1134 PE=4 SV=1
MM1 pKa = 7.16LTSLYY6 pKa = 10.65NLASSALKK14 pKa = 10.06NAQVSINNASNNIANADD31 pKa = 3.62TEE33 pKa = 4.93GYY35 pKa = 10.01QRR37 pKa = 11.84TEE39 pKa = 3.35AAYY42 pKa = 6.58EE43 pKa = 3.88TSYY46 pKa = 11.11SISIYY51 pKa = 10.08GLSVGTGANITSIISLKK68 pKa = 10.63DD69 pKa = 3.19QFVEE73 pKa = 4.21AQYY76 pKa = 11.61LDD78 pKa = 3.45ASADD82 pKa = 3.8LNRR85 pKa = 11.84EE86 pKa = 4.18SAALTYY92 pKa = 10.8LNQLDD97 pKa = 4.07SLLNQAEE104 pKa = 4.56GGLSEE109 pKa = 4.34TLTDD113 pKa = 6.78FFDD116 pKa = 3.24AWNALTTDD124 pKa = 4.23PDD126 pKa = 3.92SLSARR131 pKa = 11.84EE132 pKa = 4.17DD133 pKa = 3.49LLGIAEE139 pKa = 4.33TLVYY143 pKa = 10.66ALNSTADD150 pKa = 3.51GLEE153 pKa = 4.06TMEE156 pKa = 4.53AAINTEE162 pKa = 3.6IEE164 pKa = 4.38DD165 pKa = 4.03AVSSANQLIADD176 pKa = 4.06IAVANASIAASPDD189 pKa = 3.32NNQLISDD196 pKa = 4.42RR197 pKa = 11.84DD198 pKa = 3.45QMIRR202 pKa = 11.84EE203 pKa = 4.31LNAIIGVQTIEE214 pKa = 4.17QEE216 pKa = 4.01NGMVTILTDD225 pKa = 3.21EE226 pKa = 5.19GYY228 pKa = 10.93SLVDD232 pKa = 3.24GTEE235 pKa = 4.04THH237 pKa = 6.19SLVYY241 pKa = 10.67GDD243 pKa = 4.32ATATSALVRR252 pKa = 11.84ASDD255 pKa = 3.83YY256 pKa = 11.31DD257 pKa = 3.72GEE259 pKa = 4.31LAYY262 pKa = 10.9SGTSSEE268 pKa = 4.13EE269 pKa = 3.72VLLKK273 pKa = 10.38FVSSGADD280 pKa = 3.04GTAQFKK286 pKa = 10.94ASLDD290 pKa = 3.71GGKK293 pKa = 7.63TWITDD298 pKa = 3.63EE299 pKa = 4.35NGEE302 pKa = 4.26TMLYY306 pKa = 9.93IAGDD310 pKa = 3.37EE311 pKa = 4.06AAPVEE316 pKa = 4.17IAGIEE321 pKa = 3.69IWFEE325 pKa = 4.02GGSADD330 pKa = 4.09HH331 pKa = 6.97AVGDD335 pKa = 4.35TYY337 pKa = 11.1TIVPKK342 pKa = 10.75SGLYY346 pKa = 8.9WEE348 pKa = 5.12KK349 pKa = 11.32SDD351 pKa = 4.16GSLVNITPMTDD362 pKa = 2.92SSGTAVNGRR371 pKa = 11.84TSGGSLAGLFNVRR384 pKa = 11.84DD385 pKa = 3.97DD386 pKa = 4.11VVTPTLDD393 pKa = 3.41SLNDD397 pKa = 3.68LSEE400 pKa = 4.66SIIWEE405 pKa = 4.12VNAVHH410 pKa = 5.8STGAGLDD417 pKa = 3.34HH418 pKa = 7.22HH419 pKa = 6.46EE420 pKa = 4.63TLTGSYY426 pKa = 8.53TVEE429 pKa = 4.07DD430 pKa = 3.65SGALLSNSGLHH441 pKa = 6.26FADD444 pKa = 4.02NVTAGDD450 pKa = 4.29FSLVTYY456 pKa = 10.63DD457 pKa = 3.18SDD459 pKa = 4.8GNVSTSAIISFDD471 pKa = 3.71PATDD475 pKa = 3.69SLDD478 pKa = 3.91DD479 pKa = 4.91LMAEE483 pKa = 4.17INTAFGGEE491 pKa = 4.04LTASINASGQFVLSAGSDD509 pKa = 3.3TGFEE513 pKa = 4.14IASDD517 pKa = 3.86STGLMAALGVNTFFSGIDD535 pKa = 3.38ADD537 pKa = 4.51TIAIASAVAEE547 pKa = 4.1DD548 pKa = 3.81TSRR551 pKa = 11.84INAGAVGSDD560 pKa = 3.38GLVSSGNNDD569 pKa = 2.77IATLLAALSDD579 pKa = 4.03SKK581 pKa = 11.71VGVGDD586 pKa = 4.05KK587 pKa = 9.46NTSLTSYY594 pKa = 9.97LASLVAGVGAAASSAEE610 pKa = 4.13LKK612 pKa = 9.4LTYY615 pKa = 10.44AQTSVDD621 pKa = 3.78YY622 pKa = 10.64YY623 pKa = 10.45YY624 pKa = 10.51QQQASASEE632 pKa = 4.29VNVDD636 pKa = 3.58EE637 pKa = 5.11EE638 pKa = 5.41LINLLKK644 pKa = 10.48YY645 pKa = 9.84QQAFKK650 pKa = 10.65AAAEE654 pKa = 4.12IMTVTRR660 pKa = 11.84TMMDD664 pKa = 3.16TVLDD668 pKa = 3.86IVV670 pKa = 3.82
MM1 pKa = 7.16LTSLYY6 pKa = 10.65NLASSALKK14 pKa = 10.06NAQVSINNASNNIANADD31 pKa = 3.62TEE33 pKa = 4.93GYY35 pKa = 10.01QRR37 pKa = 11.84TEE39 pKa = 3.35AAYY42 pKa = 6.58EE43 pKa = 3.88TSYY46 pKa = 11.11SISIYY51 pKa = 10.08GLSVGTGANITSIISLKK68 pKa = 10.63DD69 pKa = 3.19QFVEE73 pKa = 4.21AQYY76 pKa = 11.61LDD78 pKa = 3.45ASADD82 pKa = 3.8LNRR85 pKa = 11.84EE86 pKa = 4.18SAALTYY92 pKa = 10.8LNQLDD97 pKa = 4.07SLLNQAEE104 pKa = 4.56GGLSEE109 pKa = 4.34TLTDD113 pKa = 6.78FFDD116 pKa = 3.24AWNALTTDD124 pKa = 4.23PDD126 pKa = 3.92SLSARR131 pKa = 11.84EE132 pKa = 4.17DD133 pKa = 3.49LLGIAEE139 pKa = 4.33TLVYY143 pKa = 10.66ALNSTADD150 pKa = 3.51GLEE153 pKa = 4.06TMEE156 pKa = 4.53AAINTEE162 pKa = 3.6IEE164 pKa = 4.38DD165 pKa = 4.03AVSSANQLIADD176 pKa = 4.06IAVANASIAASPDD189 pKa = 3.32NNQLISDD196 pKa = 4.42RR197 pKa = 11.84DD198 pKa = 3.45QMIRR202 pKa = 11.84EE203 pKa = 4.31LNAIIGVQTIEE214 pKa = 4.17QEE216 pKa = 4.01NGMVTILTDD225 pKa = 3.21EE226 pKa = 5.19GYY228 pKa = 10.93SLVDD232 pKa = 3.24GTEE235 pKa = 4.04THH237 pKa = 6.19SLVYY241 pKa = 10.67GDD243 pKa = 4.32ATATSALVRR252 pKa = 11.84ASDD255 pKa = 3.83YY256 pKa = 11.31DD257 pKa = 3.72GEE259 pKa = 4.31LAYY262 pKa = 10.9SGTSSEE268 pKa = 4.13EE269 pKa = 3.72VLLKK273 pKa = 10.38FVSSGADD280 pKa = 3.04GTAQFKK286 pKa = 10.94ASLDD290 pKa = 3.71GGKK293 pKa = 7.63TWITDD298 pKa = 3.63EE299 pKa = 4.35NGEE302 pKa = 4.26TMLYY306 pKa = 9.93IAGDD310 pKa = 3.37EE311 pKa = 4.06AAPVEE316 pKa = 4.17IAGIEE321 pKa = 3.69IWFEE325 pKa = 4.02GGSADD330 pKa = 4.09HH331 pKa = 6.97AVGDD335 pKa = 4.35TYY337 pKa = 11.1TIVPKK342 pKa = 10.75SGLYY346 pKa = 8.9WEE348 pKa = 5.12KK349 pKa = 11.32SDD351 pKa = 4.16GSLVNITPMTDD362 pKa = 2.92SSGTAVNGRR371 pKa = 11.84TSGGSLAGLFNVRR384 pKa = 11.84DD385 pKa = 3.97DD386 pKa = 4.11VVTPTLDD393 pKa = 3.41SLNDD397 pKa = 3.68LSEE400 pKa = 4.66SIIWEE405 pKa = 4.12VNAVHH410 pKa = 5.8STGAGLDD417 pKa = 3.34HH418 pKa = 7.22HH419 pKa = 6.46EE420 pKa = 4.63TLTGSYY426 pKa = 8.53TVEE429 pKa = 4.07DD430 pKa = 3.65SGALLSNSGLHH441 pKa = 6.26FADD444 pKa = 4.02NVTAGDD450 pKa = 4.29FSLVTYY456 pKa = 10.63DD457 pKa = 3.18SDD459 pKa = 4.8GNVSTSAIISFDD471 pKa = 3.71PATDD475 pKa = 3.69SLDD478 pKa = 3.91DD479 pKa = 4.91LMAEE483 pKa = 4.17INTAFGGEE491 pKa = 4.04LTASINASGQFVLSAGSDD509 pKa = 3.3TGFEE513 pKa = 4.14IASDD517 pKa = 3.86STGLMAALGVNTFFSGIDD535 pKa = 3.38ADD537 pKa = 4.51TIAIASAVAEE547 pKa = 4.1DD548 pKa = 3.81TSRR551 pKa = 11.84INAGAVGSDD560 pKa = 3.38GLVSSGNNDD569 pKa = 2.77IATLLAALSDD579 pKa = 4.03SKK581 pKa = 11.71VGVGDD586 pKa = 4.05KK587 pKa = 9.46NTSLTSYY594 pKa = 9.97LASLVAGVGAAASSAEE610 pKa = 4.13LKK612 pKa = 9.4LTYY615 pKa = 10.44AQTSVDD621 pKa = 3.78YY622 pKa = 10.64YY623 pKa = 10.45YY624 pKa = 10.51QQQASASEE632 pKa = 4.29VNVDD636 pKa = 3.58EE637 pKa = 5.11EE638 pKa = 5.41LINLLKK644 pKa = 10.48YY645 pKa = 9.84QQAFKK650 pKa = 10.65AAAEE654 pKa = 4.12IMTVTRR660 pKa = 11.84TMMDD664 pKa = 3.16TVLDD668 pKa = 3.86IVV670 pKa = 3.82
Molecular weight: 69.8 kDa
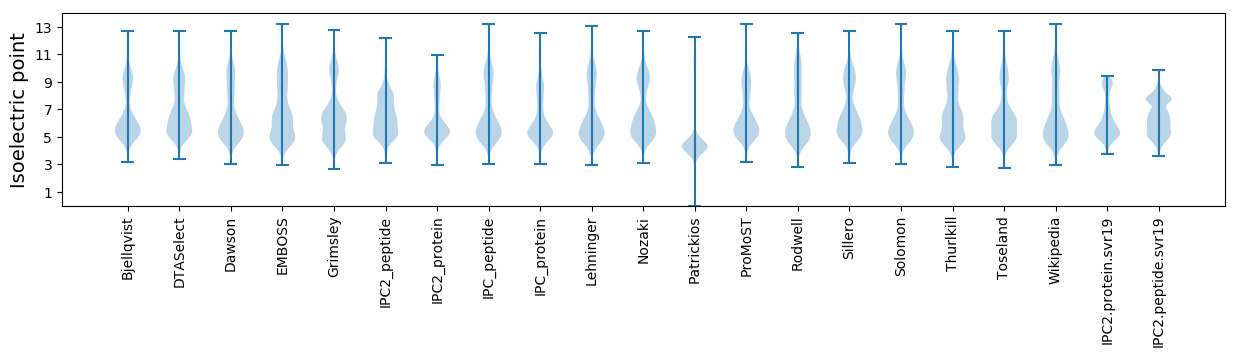
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6VRP6|E6VRP6_PSEA9 Heat shock protein Hsp20 OS=Pseudodesulfovibrio aespoeensis (strain ATCC 700646 / DSM 10631 / Aspo-2) OX=643562 GN=Daes_3191 PE=3 SV=1
MM1 pKa = 7.7PKK3 pKa = 9.95IKK5 pKa = 9.47TRR7 pKa = 11.84RR8 pKa = 11.84AAAKK12 pKa = 9.63RR13 pKa = 11.84FSKK16 pKa = 9.46TASGKK21 pKa = 9.4FKK23 pKa = 10.67RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 9.07NLRR30 pKa = 11.84HH31 pKa = 6.24ILTKK35 pKa = 10.67KK36 pKa = 7.68NAKK39 pKa = 9.22RR40 pKa = 11.84KK41 pKa = 9.18RR42 pKa = 11.84RR43 pKa = 11.84LGQSTLVDD51 pKa = 3.93STNMKK56 pKa = 9.88AVRR59 pKa = 11.84RR60 pKa = 11.84QLPNGG65 pKa = 3.51
MM1 pKa = 7.7PKK3 pKa = 9.95IKK5 pKa = 9.47TRR7 pKa = 11.84RR8 pKa = 11.84AAAKK12 pKa = 9.63RR13 pKa = 11.84FSKK16 pKa = 9.46TASGKK21 pKa = 9.4FKK23 pKa = 10.67RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 9.07NLRR30 pKa = 11.84HH31 pKa = 6.24ILTKK35 pKa = 10.67KK36 pKa = 7.68NAKK39 pKa = 9.22RR40 pKa = 11.84KK41 pKa = 9.18RR42 pKa = 11.84RR43 pKa = 11.84LGQSTLVDD51 pKa = 3.93STNMKK56 pKa = 9.88AVRR59 pKa = 11.84RR60 pKa = 11.84QLPNGG65 pKa = 3.51
Molecular weight: 7.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066561 |
31 |
3450 |
326.3 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.298 ± 0.057 | 1.314 ± 0.019 |
5.837 ± 0.04 | 6.127 ± 0.036 |
3.998 ± 0.03 | 8.346 ± 0.047 |
2.106 ± 0.022 | 5.251 ± 0.035 |
4.061 ± 0.04 | 10.429 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.893 ± 0.019 | 2.888 ± 0.029 |
4.778 ± 0.032 | 3.124 ± 0.023 |
6.605 ± 0.04 | 5.419 ± 0.036 |
5.371 ± 0.045 | 7.439 ± 0.038 |
1.167 ± 0.016 | 2.55 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
