
Blochmannia floridanus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Enterobacteriaceae incertae sedis; ant, tsetse, mealybug, aphid, etc. endosymbionts; ant endosymbionts; Candidatus Blochmannia
Average proteome isoelectric point is 7.86
Get precalculated fractions of proteins
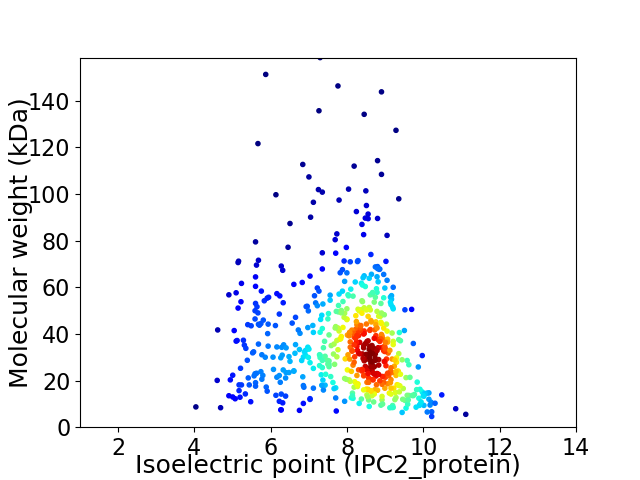
Virtual 2D-PAGE plot for 583 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
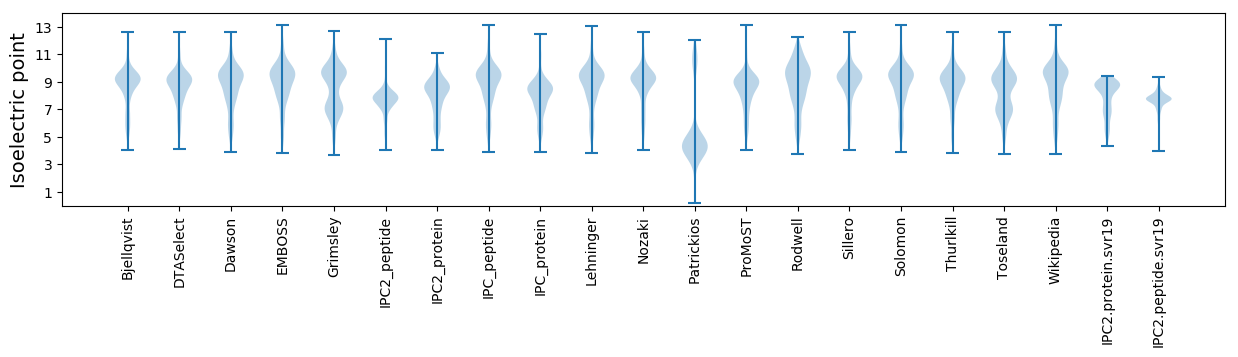
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7VR24|Q7VR24_BLOFL DNA polymerase III subunit delta' OS=Blochmannia floridanus OX=203907 GN=holB PE=4 SV=1
MM1 pKa = 7.48SIVEE5 pKa = 4.53EE6 pKa = 4.11KK7 pKa = 10.77VKK9 pKa = 10.18TIIAEE14 pKa = 3.96QLGVKK19 pKa = 9.13QEE21 pKa = 4.05EE22 pKa = 4.95VVNHH26 pKa = 6.54ASFVDD31 pKa = 3.91DD32 pKa = 5.57LGADD36 pKa = 3.42SLDD39 pKa = 3.64TVEE42 pKa = 5.9LVMALEE48 pKa = 4.32EE49 pKa = 4.24EE50 pKa = 4.31FDD52 pKa = 3.79IEE54 pKa = 4.81IPDD57 pKa = 3.67EE58 pKa = 4.11EE59 pKa = 4.53AEE61 pKa = 5.03KK62 pKa = 9.69ITTVQAAIDD71 pKa = 4.51FIHH74 pKa = 6.61TNQKK78 pKa = 10.13
MM1 pKa = 7.48SIVEE5 pKa = 4.53EE6 pKa = 4.11KK7 pKa = 10.77VKK9 pKa = 10.18TIIAEE14 pKa = 3.96QLGVKK19 pKa = 9.13QEE21 pKa = 4.05EE22 pKa = 4.95VVNHH26 pKa = 6.54ASFVDD31 pKa = 3.91DD32 pKa = 5.57LGADD36 pKa = 3.42SLDD39 pKa = 3.64TVEE42 pKa = 5.9LVMALEE48 pKa = 4.32EE49 pKa = 4.24EE50 pKa = 4.31FDD52 pKa = 3.79IEE54 pKa = 4.81IPDD57 pKa = 3.67EE58 pKa = 4.11EE59 pKa = 4.53AEE61 pKa = 5.03KK62 pKa = 9.69ITTVQAAIDD71 pKa = 4.51FIHH74 pKa = 6.61TNQKK78 pKa = 10.13
Molecular weight: 8.7 kDa
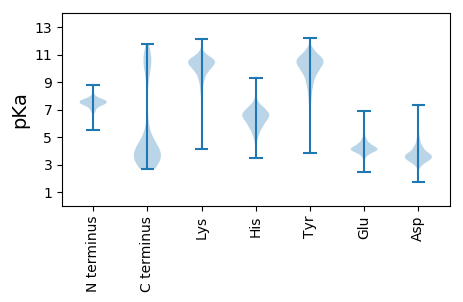
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q7VQV3|MNME_BLOFL tRNA modification GTPase MnmE OS=Blochmannia floridanus OX=203907 GN=mnmE PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.14QPSVLKK11 pKa = 10.65RR12 pKa = 11.84NRR14 pKa = 11.84THH16 pKa = 7.22GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTQNGRR28 pKa = 11.84QILSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.6GRR39 pKa = 11.84IRR41 pKa = 11.84LIVSSS46 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.14QPSVLKK11 pKa = 10.65RR12 pKa = 11.84NRR14 pKa = 11.84THH16 pKa = 7.22GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTQNGRR28 pKa = 11.84QILSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.6GRR39 pKa = 11.84IRR41 pKa = 11.84LIVSSS46 pKa = 4.03
Molecular weight: 5.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
195064 |
38 |
1420 |
334.6 |
38.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.566 ± 0.081 | 1.578 ± 0.037 |
4.585 ± 0.068 | 4.439 ± 0.075 |
4.453 ± 0.076 | 5.7 ± 0.096 |
2.581 ± 0.063 | 11.407 ± 0.118 |
7.168 ± 0.081 | 10.051 ± 0.102 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.508 ± 0.047 | 6.889 ± 0.108 |
3.18 ± 0.048 | 3.9 ± 0.067 |
4.082 ± 0.074 | 7.029 ± 0.064 |
4.864 ± 0.068 | 5.751 ± 0.12 |
1.037 ± 0.034 | 4.232 ± 0.069 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
