
Cyclospora cayetanensis
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Conoidasida; Coccidia; Eucoccidiorida; Eimeriorina; Eimeriidae; Cyclospora
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
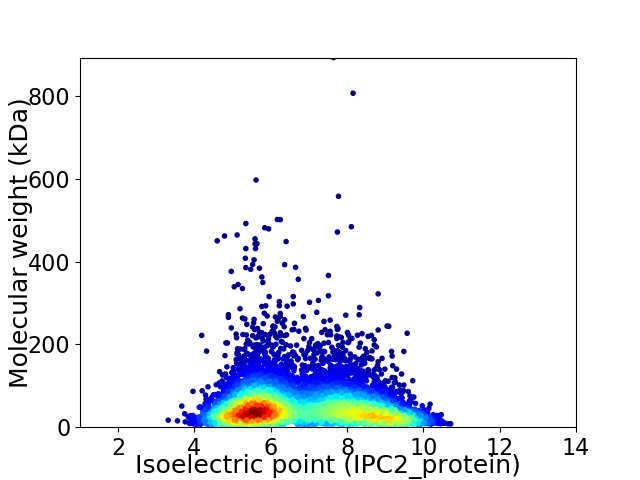
Virtual 2D-PAGE plot for 7151 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D3CS24|A0A1D3CS24_9EIME Uncharacterized protein OS=Cyclospora cayetanensis OX=88456 GN=cyc_07968 PE=4 SV=1
MM1 pKa = 7.44TPPLRR6 pKa = 11.84GSPTGRR12 pKa = 11.84LSAAAAAIATAAMSALLLSPSAKK35 pKa = 9.86AQEE38 pKa = 4.58PYY40 pKa = 10.53GVPSEE45 pKa = 4.87ASWAPASASSTLSYY59 pKa = 10.81SPSAFSASASPSAYY73 pKa = 9.3ISPSLSFSASSSYY86 pKa = 11.11QYY88 pKa = 11.13TNYY91 pKa = 10.09PEE93 pKa = 4.11YY94 pKa = 10.08QGSGGGPQVASEE106 pKa = 4.19LSGLGYY112 pKa = 8.37FTASLDD118 pKa = 3.42TRR120 pKa = 11.84AISAGQYY127 pKa = 9.08SASPEE132 pKa = 4.23TFSVSPSSSIPAFDD146 pKa = 3.74SFTPYY151 pKa = 10.71APPAAPSFAPSAAPPFAPSAAPSSAAAYY179 pKa = 9.52QDD181 pKa = 3.79AGIATTEE188 pKa = 3.98ATATAASAYY197 pKa = 8.26EE198 pKa = 4.34AYY200 pKa = 10.45ASGGLTASSGSPYY213 pKa = 10.03TPASNPVSPSYY224 pKa = 10.7FDD226 pKa = 3.69EE227 pKa = 5.75GPTEE231 pKa = 4.12MAPLPSPSMPVAGVAEE247 pKa = 4.7AAAEE251 pKa = 4.1MQTAAVAAVAADD263 pKa = 3.92TTATADD269 pKa = 3.39MAEE272 pKa = 4.09AAVAAEE278 pKa = 4.06AAAAALEE285 pKa = 4.09ALQEE289 pKa = 4.4TLTSPTQHH297 pKa = 6.71HH298 pKa = 6.53EE299 pKa = 3.98ATTEE303 pKa = 3.77AVANTEE309 pKa = 4.13QVDD312 pKa = 3.43SHH314 pKa = 6.72AAEE317 pKa = 4.01TVAAQGAPEE326 pKa = 4.85HH327 pKa = 6.59PPSVQVSPTADD338 pKa = 3.18SSPDD342 pKa = 3.45TVSAEE347 pKa = 4.06TTAARR352 pKa = 11.84QPTEE356 pKa = 3.94SSEE359 pKa = 3.96AAQQEE364 pKa = 4.66GAEE367 pKa = 4.36SARR370 pKa = 11.84VQDD373 pKa = 3.65EE374 pKa = 4.11ASAVALTEE382 pKa = 3.81PTAAQTPSEE391 pKa = 4.52GQQHH395 pKa = 5.58TPVEE399 pKa = 4.49EE400 pKa = 3.95QTQQQGEE407 pKa = 4.34TAEE410 pKa = 4.4GLLTAAAGDD419 pKa = 4.19TYY421 pKa = 11.63VEE423 pKa = 4.46TPAAASTTQSAEE435 pKa = 3.57VDD437 pKa = 3.51AGGHH441 pKa = 4.29TTLAEE446 pKa = 4.2GTTALPSEE454 pKa = 4.44AVMPLAEE461 pKa = 4.2AAEE464 pKa = 4.26AQHH467 pKa = 6.68ASSHH471 pKa = 6.52PSQSATGEE479 pKa = 3.93ASEE482 pKa = 5.31DD483 pKa = 3.44QQQQSQQPNEE493 pKa = 4.05PPQLQQATEE502 pKa = 3.69THH504 pKa = 6.16AAYY507 pKa = 9.59EE508 pKa = 4.31SLGALPNAHH517 pKa = 6.35PQEE520 pKa = 4.52EE521 pKa = 5.3GSTEE525 pKa = 4.09GAPTGAATPPSSLSAAVDD543 pKa = 3.61EE544 pKa = 4.85AVMFAAAINAPSSAPAEE561 pKa = 4.23AGSADD566 pKa = 3.71EE567 pKa = 5.52AEE569 pKa = 4.5SQAAATQGGMLPEE582 pKa = 5.14SYY584 pKa = 10.53PEE586 pKa = 4.58GPTLEE591 pKa = 4.4TPLEE595 pKa = 4.02EE596 pKa = 4.1MMMEE600 pKa = 4.54HH601 pKa = 6.75AQNNPAAATQAEE613 pKa = 4.64EE614 pKa = 4.97DD615 pKa = 3.55SQGPHH620 pKa = 6.5RR621 pKa = 11.84EE622 pKa = 4.08SEE624 pKa = 4.27AAGAAPAADD633 pKa = 3.94GQLDD637 pKa = 3.67AAASEE642 pKa = 4.29PTQRR646 pKa = 11.84GALDD650 pKa = 3.49TLTGVTQQAIAFGSHH665 pKa = 7.12PEE667 pKa = 4.12SEE669 pKa = 4.28TDD671 pKa = 3.19EE672 pKa = 4.38TAEE675 pKa = 4.31SQQQQHH681 pKa = 7.1PGQSQQQRR689 pKa = 11.84GQGTLEE695 pKa = 4.04QEE697 pKa = 4.39ASAQQHH703 pKa = 6.08EE704 pKa = 4.73IVIRR708 pKa = 11.84LLTGDD713 pKa = 4.58KK714 pKa = 10.53EE715 pKa = 4.23DD716 pKa = 4.21LAASLSRR723 pKa = 11.84LAEE726 pKa = 4.42TIRR729 pKa = 11.84HH730 pKa = 5.79RR731 pKa = 11.84EE732 pKa = 3.66RR733 pKa = 11.84LANPHH738 pKa = 6.41EE739 pKa = 4.28EE740 pKa = 4.04LATVEE745 pKa = 4.13GTEE748 pKa = 4.24QQEE751 pKa = 4.38QQQTPADD758 pKa = 3.55ATEE761 pKa = 4.05NAEE764 pKa = 4.38GVAAPLKK771 pKa = 10.39PGTPQEE777 pKa = 4.26EE778 pKa = 4.46EE779 pKa = 3.64QDD781 pKa = 3.6QSGEE785 pKa = 4.12EE786 pKa = 4.11TPDD789 pKa = 3.08RR790 pKa = 11.84SRR792 pKa = 11.84QYY794 pKa = 10.39IFQSVSQQQQQDD806 pKa = 3.33LPAEE810 pKa = 4.32VEE812 pKa = 3.96ALQQVLIQQQLQQTHH827 pKa = 4.47QQVLLEE833 pKa = 4.21EE834 pKa = 4.28EE835 pKa = 4.67LNGSVEE841 pKa = 4.25VPDD844 pKa = 3.51IEE846 pKa = 6.46AYY848 pKa = 9.74VQQ850 pKa = 3.24
MM1 pKa = 7.44TPPLRR6 pKa = 11.84GSPTGRR12 pKa = 11.84LSAAAAAIATAAMSALLLSPSAKK35 pKa = 9.86AQEE38 pKa = 4.58PYY40 pKa = 10.53GVPSEE45 pKa = 4.87ASWAPASASSTLSYY59 pKa = 10.81SPSAFSASASPSAYY73 pKa = 9.3ISPSLSFSASSSYY86 pKa = 11.11QYY88 pKa = 11.13TNYY91 pKa = 10.09PEE93 pKa = 4.11YY94 pKa = 10.08QGSGGGPQVASEE106 pKa = 4.19LSGLGYY112 pKa = 8.37FTASLDD118 pKa = 3.42TRR120 pKa = 11.84AISAGQYY127 pKa = 9.08SASPEE132 pKa = 4.23TFSVSPSSSIPAFDD146 pKa = 3.74SFTPYY151 pKa = 10.71APPAAPSFAPSAAPPFAPSAAPSSAAAYY179 pKa = 9.52QDD181 pKa = 3.79AGIATTEE188 pKa = 3.98ATATAASAYY197 pKa = 8.26EE198 pKa = 4.34AYY200 pKa = 10.45ASGGLTASSGSPYY213 pKa = 10.03TPASNPVSPSYY224 pKa = 10.7FDD226 pKa = 3.69EE227 pKa = 5.75GPTEE231 pKa = 4.12MAPLPSPSMPVAGVAEE247 pKa = 4.7AAAEE251 pKa = 4.1MQTAAVAAVAADD263 pKa = 3.92TTATADD269 pKa = 3.39MAEE272 pKa = 4.09AAVAAEE278 pKa = 4.06AAAAALEE285 pKa = 4.09ALQEE289 pKa = 4.4TLTSPTQHH297 pKa = 6.71HH298 pKa = 6.53EE299 pKa = 3.98ATTEE303 pKa = 3.77AVANTEE309 pKa = 4.13QVDD312 pKa = 3.43SHH314 pKa = 6.72AAEE317 pKa = 4.01TVAAQGAPEE326 pKa = 4.85HH327 pKa = 6.59PPSVQVSPTADD338 pKa = 3.18SSPDD342 pKa = 3.45TVSAEE347 pKa = 4.06TTAARR352 pKa = 11.84QPTEE356 pKa = 3.94SSEE359 pKa = 3.96AAQQEE364 pKa = 4.66GAEE367 pKa = 4.36SARR370 pKa = 11.84VQDD373 pKa = 3.65EE374 pKa = 4.11ASAVALTEE382 pKa = 3.81PTAAQTPSEE391 pKa = 4.52GQQHH395 pKa = 5.58TPVEE399 pKa = 4.49EE400 pKa = 3.95QTQQQGEE407 pKa = 4.34TAEE410 pKa = 4.4GLLTAAAGDD419 pKa = 4.19TYY421 pKa = 11.63VEE423 pKa = 4.46TPAAASTTQSAEE435 pKa = 3.57VDD437 pKa = 3.51AGGHH441 pKa = 4.29TTLAEE446 pKa = 4.2GTTALPSEE454 pKa = 4.44AVMPLAEE461 pKa = 4.2AAEE464 pKa = 4.26AQHH467 pKa = 6.68ASSHH471 pKa = 6.52PSQSATGEE479 pKa = 3.93ASEE482 pKa = 5.31DD483 pKa = 3.44QQQQSQQPNEE493 pKa = 4.05PPQLQQATEE502 pKa = 3.69THH504 pKa = 6.16AAYY507 pKa = 9.59EE508 pKa = 4.31SLGALPNAHH517 pKa = 6.35PQEE520 pKa = 4.52EE521 pKa = 5.3GSTEE525 pKa = 4.09GAPTGAATPPSSLSAAVDD543 pKa = 3.61EE544 pKa = 4.85AVMFAAAINAPSSAPAEE561 pKa = 4.23AGSADD566 pKa = 3.71EE567 pKa = 5.52AEE569 pKa = 4.5SQAAATQGGMLPEE582 pKa = 5.14SYY584 pKa = 10.53PEE586 pKa = 4.58GPTLEE591 pKa = 4.4TPLEE595 pKa = 4.02EE596 pKa = 4.1MMMEE600 pKa = 4.54HH601 pKa = 6.75AQNNPAAATQAEE613 pKa = 4.64EE614 pKa = 4.97DD615 pKa = 3.55SQGPHH620 pKa = 6.5RR621 pKa = 11.84EE622 pKa = 4.08SEE624 pKa = 4.27AAGAAPAADD633 pKa = 3.94GQLDD637 pKa = 3.67AAASEE642 pKa = 4.29PTQRR646 pKa = 11.84GALDD650 pKa = 3.49TLTGVTQQAIAFGSHH665 pKa = 7.12PEE667 pKa = 4.12SEE669 pKa = 4.28TDD671 pKa = 3.19EE672 pKa = 4.38TAEE675 pKa = 4.31SQQQQHH681 pKa = 7.1PGQSQQQRR689 pKa = 11.84GQGTLEE695 pKa = 4.04QEE697 pKa = 4.39ASAQQHH703 pKa = 6.08EE704 pKa = 4.73IVIRR708 pKa = 11.84LLTGDD713 pKa = 4.58KK714 pKa = 10.53EE715 pKa = 4.23DD716 pKa = 4.21LAASLSRR723 pKa = 11.84LAEE726 pKa = 4.42TIRR729 pKa = 11.84HH730 pKa = 5.79RR731 pKa = 11.84EE732 pKa = 3.66RR733 pKa = 11.84LANPHH738 pKa = 6.41EE739 pKa = 4.28EE740 pKa = 4.04LATVEE745 pKa = 4.13GTEE748 pKa = 4.24QQEE751 pKa = 4.38QQQTPADD758 pKa = 3.55ATEE761 pKa = 4.05NAEE764 pKa = 4.38GVAAPLKK771 pKa = 10.39PGTPQEE777 pKa = 4.26EE778 pKa = 4.46EE779 pKa = 3.64QDD781 pKa = 3.6QSGEE785 pKa = 4.12EE786 pKa = 4.11TPDD789 pKa = 3.08RR790 pKa = 11.84SRR792 pKa = 11.84QYY794 pKa = 10.39IFQSVSQQQQQDD806 pKa = 3.33LPAEE810 pKa = 4.32VEE812 pKa = 3.96ALQQVLIQQQLQQTHH827 pKa = 4.47QQVLLEE833 pKa = 4.21EE834 pKa = 4.28EE835 pKa = 4.67LNGSVEE841 pKa = 4.25VPDD844 pKa = 3.51IEE846 pKa = 6.46AYY848 pKa = 9.74VQQ850 pKa = 3.24
Molecular weight: 86.73 kDa
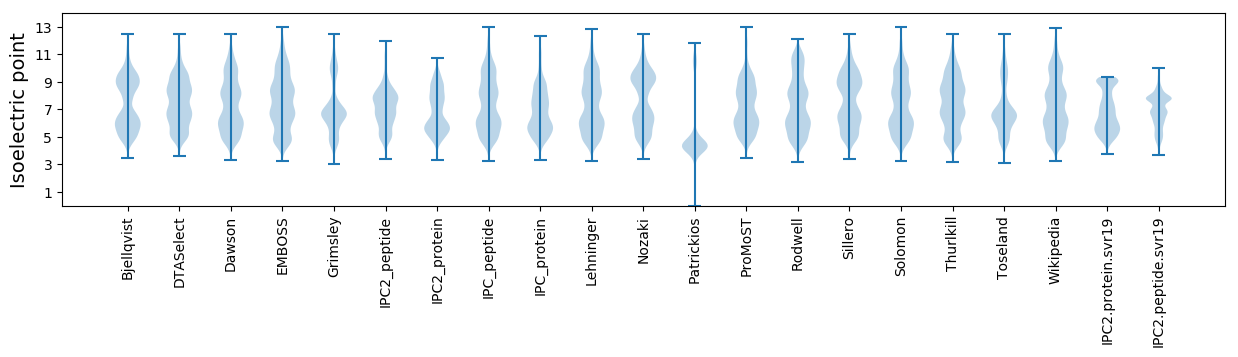
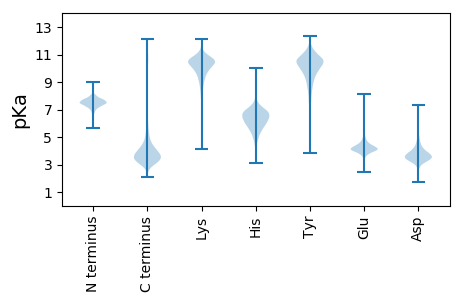
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D3CZ42|A0A1D3CZ42_9EIME Vacuolar protein sorting OS=Cyclospora cayetanensis OX=88456 GN=cyc_05819 PE=4 SV=1
MM1 pKa = 7.95RR2 pKa = 11.84GHH4 pKa = 6.73CTPGDD9 pKa = 3.17WAYY12 pKa = 11.56VMEE15 pKa = 4.66VSDD18 pKa = 5.1PEE20 pKa = 3.95QQLRR24 pKa = 11.84RR25 pKa = 11.84FMRR28 pKa = 11.84QAFSAEE34 pKa = 4.08GGPLLNVLSRR44 pKa = 11.84DD45 pKa = 3.96RR46 pKa = 11.84IQLWGSGRR54 pKa = 11.84LRR56 pKa = 11.84SPSSRR61 pKa = 11.84RR62 pKa = 11.84LAQASSRR69 pKa = 11.84FCKK72 pKa = 10.48GLFSRR77 pKa = 11.84VPPRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84EE84 pKa = 3.85GLRR87 pKa = 11.84DD88 pKa = 3.67SNASSSLPAPHH99 pKa = 7.29EE100 pKa = 4.54SPLMGPPKK108 pKa = 9.95PPPAPQDD115 pKa = 3.75SNMMRR120 pKa = 11.84ASVHH124 pKa = 5.88ARR126 pKa = 11.84CLQDD130 pKa = 3.83FYY132 pKa = 11.76FRR134 pKa = 11.84LEE136 pKa = 3.91EE137 pKa = 4.11EE138 pKa = 4.2KK139 pKa = 11.09APGYY143 pKa = 8.14VLCICVGPPSRR154 pKa = 11.84ALKK157 pKa = 9.91EE158 pKa = 4.42YY159 pKa = 9.15KK160 pKa = 9.89SCMPRR165 pKa = 11.84VRR167 pKa = 11.84LRR169 pKa = 11.84GKK171 pKa = 9.99GGRR174 pKa = 11.84TGTVPP179 pKa = 3.58
MM1 pKa = 7.95RR2 pKa = 11.84GHH4 pKa = 6.73CTPGDD9 pKa = 3.17WAYY12 pKa = 11.56VMEE15 pKa = 4.66VSDD18 pKa = 5.1PEE20 pKa = 3.95QQLRR24 pKa = 11.84RR25 pKa = 11.84FMRR28 pKa = 11.84QAFSAEE34 pKa = 4.08GGPLLNVLSRR44 pKa = 11.84DD45 pKa = 3.96RR46 pKa = 11.84IQLWGSGRR54 pKa = 11.84LRR56 pKa = 11.84SPSSRR61 pKa = 11.84RR62 pKa = 11.84LAQASSRR69 pKa = 11.84FCKK72 pKa = 10.48GLFSRR77 pKa = 11.84VPPRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84EE84 pKa = 3.85GLRR87 pKa = 11.84DD88 pKa = 3.67SNASSSLPAPHH99 pKa = 7.29EE100 pKa = 4.54SPLMGPPKK108 pKa = 9.95PPPAPQDD115 pKa = 3.75SNMMRR120 pKa = 11.84ASVHH124 pKa = 5.88ARR126 pKa = 11.84CLQDD130 pKa = 3.83FYY132 pKa = 11.76FRR134 pKa = 11.84LEE136 pKa = 3.91EE137 pKa = 4.11EE138 pKa = 4.2KK139 pKa = 11.09APGYY143 pKa = 8.14VLCICVGPPSRR154 pKa = 11.84ALKK157 pKa = 9.91EE158 pKa = 4.42YY159 pKa = 9.15KK160 pKa = 9.89SCMPRR165 pKa = 11.84VRR167 pKa = 11.84LRR169 pKa = 11.84GKK171 pKa = 9.99GGRR174 pKa = 11.84TGTVPP179 pKa = 3.58
Molecular weight: 20.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3734096 |
66 |
8164 |
522.2 |
56.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.347 ± 0.045 | 2.293 ± 0.016 |
4.208 ± 0.015 | 6.733 ± 0.024 |
3.195 ± 0.016 | 7.023 ± 0.026 |
2.465 ± 0.012 | 3.064 ± 0.018 |
4.02 ± 0.023 | 10.602 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.746 ± 0.01 | 2.344 ± 0.013 |
6.221 ± 0.027 | 5.342 ± 0.03 |
6.851 ± 0.024 | 8.841 ± 0.029 |
4.904 ± 0.016 | 5.898 ± 0.021 |
1.132 ± 0.007 | 1.764 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
