
Nocardioides sp. GY 10127
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
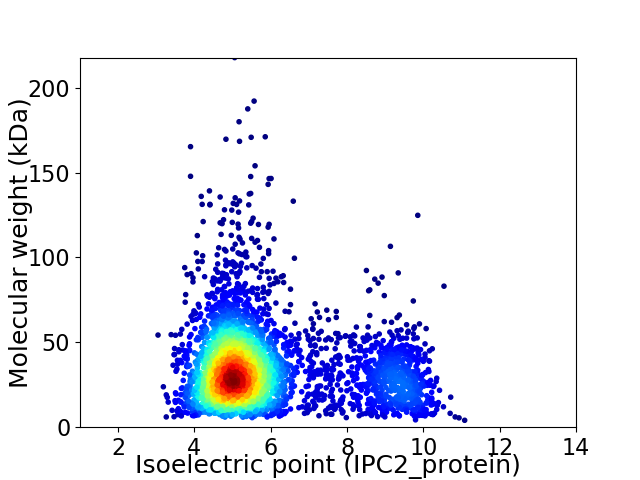
Virtual 2D-PAGE plot for 3765 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4T0USX8|A0A4T0USX8_9ACTN ATP-dependent Clp protease proteolytic subunit OS=Nocardioides sp. GY 10127 OX=2569762 GN=E8D37_11570 PE=3 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84SRR6 pKa = 11.84VAASLGVIALATTLLAACGSEE27 pKa = 4.13SSEE30 pKa = 3.84DD31 pKa = 3.64TAAEE35 pKa = 3.93TAAAGATDD43 pKa = 4.96LAAAGCPATIVIQTDD58 pKa = 3.34WNPEE62 pKa = 3.83AEE64 pKa = 4.33HH65 pKa = 5.57GHH67 pKa = 6.45LYY69 pKa = 11.17EE70 pKa = 4.69MFGDD74 pKa = 4.52DD75 pKa = 4.46ATIDD79 pKa = 3.78ADD81 pKa = 3.78NKK83 pKa = 10.24SVSGTLYY90 pKa = 11.07DD91 pKa = 3.69NGEE94 pKa = 4.19STGVKK99 pKa = 10.16LEE101 pKa = 3.86VRR103 pKa = 11.84AGGPAIGYY111 pKa = 7.08STVSAQMYY119 pKa = 7.28TDD121 pKa = 3.6KK122 pKa = 10.75SITMGYY128 pKa = 7.91VTTDD132 pKa = 3.29DD133 pKa = 4.52QIASSATTPTKK144 pKa = 10.89SFFAPLDD151 pKa = 3.85LSPLMVMWDD160 pKa = 3.28PTYY163 pKa = 10.92YY164 pKa = 10.16PDD166 pKa = 3.57VTGIDD171 pKa = 3.95DD172 pKa = 5.31LKK174 pKa = 11.32DD175 pKa = 3.64ALDD178 pKa = 3.61SSGGVWRR185 pKa = 11.84YY186 pKa = 8.08FAGSAYY192 pKa = 9.62MDD194 pKa = 3.81YY195 pKa = 10.89LEE197 pKa = 4.44SAGYY201 pKa = 9.53VSKK204 pKa = 11.33DD205 pKa = 3.64SMDD208 pKa = 3.14SSYY211 pKa = 11.7DD212 pKa = 3.29GTPANFVAAGGKK224 pKa = 9.36DD225 pKa = 3.61VQQGFASAEE234 pKa = 3.91PYY236 pKa = 10.11QYY238 pKa = 11.17EE239 pKa = 4.38NEE241 pKa = 3.99VKK243 pKa = 10.32AWGKK247 pKa = 10.18KK248 pKa = 7.63VDD250 pKa = 3.67YY251 pKa = 11.0ALISDD256 pKa = 4.49TGWDD260 pKa = 4.08PYY262 pKa = 10.94QSSLAVRR269 pKa = 11.84SADD272 pKa = 3.86FDD274 pKa = 4.38DD275 pKa = 6.81LSDD278 pKa = 4.29CLAALTPVLQRR289 pKa = 11.84AEE291 pKa = 4.16VNYY294 pKa = 10.16FADD297 pKa = 3.78PTDD300 pKa = 3.7ANNLVLEE307 pKa = 4.67AVQEE311 pKa = 4.43YY312 pKa = 7.96NTGWTYY318 pKa = 10.13SQGMADD324 pKa = 3.69YY325 pKa = 10.66SVEE328 pKa = 4.02TMLGDD333 pKa = 3.72KK334 pKa = 10.51LVSNGDD340 pKa = 2.98NDD342 pKa = 4.39YY343 pKa = 11.28IGDD346 pKa = 3.87FDD348 pKa = 6.14DD349 pKa = 5.76DD350 pKa = 4.22RR351 pKa = 11.84MSAFFDD357 pKa = 3.81KK358 pKa = 10.39ATKK361 pKa = 9.65TYY363 pKa = 10.66SALGTDD369 pKa = 3.6MASGLKK375 pKa = 10.6VSDD378 pKa = 4.57LYY380 pKa = 11.27TNEE383 pKa = 5.2FIDD386 pKa = 5.43DD387 pKa = 4.38SIGLSNN393 pKa = 4.32
MM1 pKa = 7.65RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84SRR6 pKa = 11.84VAASLGVIALATTLLAACGSEE27 pKa = 4.13SSEE30 pKa = 3.84DD31 pKa = 3.64TAAEE35 pKa = 3.93TAAAGATDD43 pKa = 4.96LAAAGCPATIVIQTDD58 pKa = 3.34WNPEE62 pKa = 3.83AEE64 pKa = 4.33HH65 pKa = 5.57GHH67 pKa = 6.45LYY69 pKa = 11.17EE70 pKa = 4.69MFGDD74 pKa = 4.52DD75 pKa = 4.46ATIDD79 pKa = 3.78ADD81 pKa = 3.78NKK83 pKa = 10.24SVSGTLYY90 pKa = 11.07DD91 pKa = 3.69NGEE94 pKa = 4.19STGVKK99 pKa = 10.16LEE101 pKa = 3.86VRR103 pKa = 11.84AGGPAIGYY111 pKa = 7.08STVSAQMYY119 pKa = 7.28TDD121 pKa = 3.6KK122 pKa = 10.75SITMGYY128 pKa = 7.91VTTDD132 pKa = 3.29DD133 pKa = 4.52QIASSATTPTKK144 pKa = 10.89SFFAPLDD151 pKa = 3.85LSPLMVMWDD160 pKa = 3.28PTYY163 pKa = 10.92YY164 pKa = 10.16PDD166 pKa = 3.57VTGIDD171 pKa = 3.95DD172 pKa = 5.31LKK174 pKa = 11.32DD175 pKa = 3.64ALDD178 pKa = 3.61SSGGVWRR185 pKa = 11.84YY186 pKa = 8.08FAGSAYY192 pKa = 9.62MDD194 pKa = 3.81YY195 pKa = 10.89LEE197 pKa = 4.44SAGYY201 pKa = 9.53VSKK204 pKa = 11.33DD205 pKa = 3.64SMDD208 pKa = 3.14SSYY211 pKa = 11.7DD212 pKa = 3.29GTPANFVAAGGKK224 pKa = 9.36DD225 pKa = 3.61VQQGFASAEE234 pKa = 3.91PYY236 pKa = 10.11QYY238 pKa = 11.17EE239 pKa = 4.38NEE241 pKa = 3.99VKK243 pKa = 10.32AWGKK247 pKa = 10.18KK248 pKa = 7.63VDD250 pKa = 3.67YY251 pKa = 11.0ALISDD256 pKa = 4.49TGWDD260 pKa = 4.08PYY262 pKa = 10.94QSSLAVRR269 pKa = 11.84SADD272 pKa = 3.86FDD274 pKa = 4.38DD275 pKa = 6.81LSDD278 pKa = 4.29CLAALTPVLQRR289 pKa = 11.84AEE291 pKa = 4.16VNYY294 pKa = 10.16FADD297 pKa = 3.78PTDD300 pKa = 3.7ANNLVLEE307 pKa = 4.67AVQEE311 pKa = 4.43YY312 pKa = 7.96NTGWTYY318 pKa = 10.13SQGMADD324 pKa = 3.69YY325 pKa = 10.66SVEE328 pKa = 4.02TMLGDD333 pKa = 3.72KK334 pKa = 10.51LVSNGDD340 pKa = 2.98NDD342 pKa = 4.39YY343 pKa = 11.28IGDD346 pKa = 3.87FDD348 pKa = 6.14DD349 pKa = 5.76DD350 pKa = 4.22RR351 pKa = 11.84MSAFFDD357 pKa = 3.81KK358 pKa = 10.39ATKK361 pKa = 9.65TYY363 pKa = 10.66SALGTDD369 pKa = 3.6MASGLKK375 pKa = 10.6VSDD378 pKa = 4.57LYY380 pKa = 11.27TNEE383 pKa = 5.2FIDD386 pKa = 5.43DD387 pKa = 4.38SIGLSNN393 pKa = 4.32
Molecular weight: 42.07 kDa
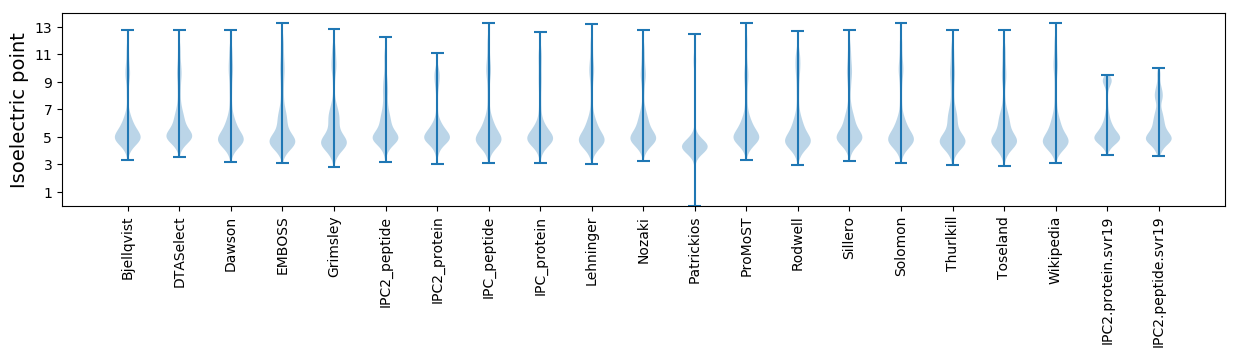
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4T0UN17|A0A4T0UN17_9ACTN GntP family transporter OS=Nocardioides sp. GY 10127 OX=2569762 GN=E8D37_15215 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1280276 |
33 |
2019 |
340.0 |
36.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.812 ± 0.065 | 0.706 ± 0.011 |
6.379 ± 0.034 | 5.698 ± 0.038 |
2.494 ± 0.024 | 9.539 ± 0.041 |
2.19 ± 0.021 | 2.641 ± 0.032 |
1.676 ± 0.031 | 10.745 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.666 ± 0.016 | 1.496 ± 0.02 |
5.697 ± 0.035 | 2.614 ± 0.021 |
7.623 ± 0.043 | 5.482 ± 0.034 |
6.352 ± 0.04 | 9.841 ± 0.042 |
1.539 ± 0.018 | 1.811 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
