
Capybara microvirus Cap1_SP_124
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.67
Get precalculated fractions of proteins
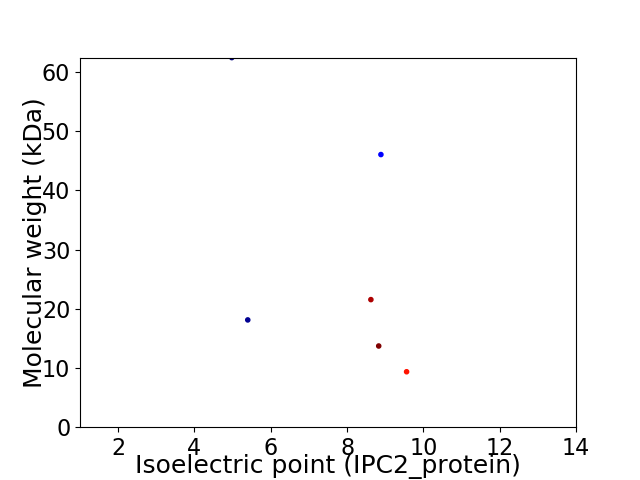
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7V9|A0A4P8W7V9_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_124 OX=2585379 PE=4 SV=1
MM1 pKa = 7.35SRR3 pKa = 11.84NEE5 pKa = 3.64LAYY8 pKa = 10.03EE9 pKa = 4.04VLDD12 pKa = 4.63TIDD15 pKa = 3.87TPRR18 pKa = 11.84STWQRR23 pKa = 11.84NFTHH27 pKa = 5.98KK28 pKa = 9.74TSFNAGKK35 pKa = 10.27LIPFYY40 pKa = 10.25IDD42 pKa = 2.91TDD44 pKa = 4.38IIPGTTIKK52 pKa = 10.78NKK54 pKa = 7.91TSIVIRR60 pKa = 11.84MSTPIAPVMDD70 pKa = 4.2NLYY73 pKa = 10.83LDD75 pKa = 4.06TYY77 pKa = 10.16YY78 pKa = 10.85FKK80 pKa = 10.92CSKK83 pKa = 9.39FWYY86 pKa = 8.05WEE88 pKa = 3.68HH89 pKa = 6.45FRR91 pKa = 11.84AQMGEE96 pKa = 4.08NNQSAWTQTIEE107 pKa = 4.47YY108 pKa = 9.45IEE110 pKa = 4.36PQIKK114 pKa = 8.49MLSMEE119 pKa = 4.1YY120 pKa = 10.41GANDD124 pKa = 3.34VACYY128 pKa = 9.56MGMPINVANLEE139 pKa = 4.18WSKK142 pKa = 11.35LPVQAYY148 pKa = 9.67IDD150 pKa = 3.21IWNNWFRR157 pKa = 11.84DD158 pKa = 3.68EE159 pKa = 4.63NLQAPITLDD168 pKa = 3.21KK169 pKa = 11.01TDD171 pKa = 4.44ADD173 pKa = 4.01LTADD177 pKa = 3.42ATINTGFGLLPVCKK191 pKa = 9.86FHH193 pKa = 8.39DD194 pKa = 4.49YY195 pKa = 8.25FTSLLPQPQKK205 pKa = 11.38GEE207 pKa = 4.09AVTMPLGLTAPVTGTVIGNGKK228 pKa = 7.37TLGLWNGTTTAGIRR242 pKa = 11.84QGIGVSGDD250 pKa = 3.22NYY252 pKa = 11.04SNGIGIASYY261 pKa = 10.98NSSNLPYY268 pKa = 8.63NTSGVSSLNNTAGLGVTTNPDD289 pKa = 2.93KK290 pKa = 11.38SGLKK294 pKa = 9.37IASNAVADD302 pKa = 4.19LTSATAATFNALRR315 pKa = 11.84LAGATQRR322 pKa = 11.84ILEE325 pKa = 4.15KK326 pKa = 10.39DD327 pKa = 3.0ARR329 pKa = 11.84FGTRR333 pKa = 11.84YY334 pKa = 10.0PEE336 pKa = 4.04FLKK339 pKa = 10.96GQYY342 pKa = 10.34GVTAADD348 pKa = 3.67EE349 pKa = 4.4CLLIPEE355 pKa = 4.23YY356 pKa = 10.85LGGKK360 pKa = 9.42RR361 pKa = 11.84IPINIEE367 pKa = 3.84QVCQTSSTDD376 pKa = 3.15STSPLGEE383 pKa = 4.06TGAMSVTADD392 pKa = 3.13INEE395 pKa = 4.73DD396 pKa = 3.43FTKK399 pKa = 10.84SFTKK403 pKa = 10.11HH404 pKa = 6.27DD405 pKa = 3.95FLIGVCCVRR414 pKa = 11.84AEE416 pKa = 3.94HH417 pKa = 6.96TYY419 pKa = 9.94QQGIPLQFKK428 pKa = 9.92RR429 pKa = 11.84KK430 pKa = 9.03RR431 pKa = 11.84RR432 pKa = 11.84LDD434 pKa = 3.49TYY436 pKa = 10.51HH437 pKa = 7.55PSLAHH442 pKa = 6.92IGNQPVYY449 pKa = 10.03NFEE452 pKa = 4.34IFAQGNSTDD461 pKa = 4.19DD462 pKa = 3.11EE463 pKa = 4.97VMGYY467 pKa = 9.92KK468 pKa = 9.81EE469 pKa = 3.85AWQEE473 pKa = 3.68YY474 pKa = 9.17LYY476 pKa = 10.9KK477 pKa = 10.26PNRR480 pKa = 11.84ISGQMLSTYY489 pKa = 7.29TQSLDD494 pKa = 2.93VWHH497 pKa = 6.74YY498 pKa = 10.96GDD500 pKa = 5.8DD501 pKa = 4.11YY502 pKa = 12.05VNLPVLSSEE511 pKa = 4.47WIVEE515 pKa = 3.72PTEE518 pKa = 4.26YY519 pKa = 9.51IDD521 pKa = 3.24RR522 pKa = 11.84TLAVSSSVSNQFWADD537 pKa = 2.99IYY539 pKa = 10.83IEE541 pKa = 4.41QIVSAPIPINRR552 pKa = 11.84VPGLIDD558 pKa = 3.56HH559 pKa = 6.51YY560 pKa = 11.72
MM1 pKa = 7.35SRR3 pKa = 11.84NEE5 pKa = 3.64LAYY8 pKa = 10.03EE9 pKa = 4.04VLDD12 pKa = 4.63TIDD15 pKa = 3.87TPRR18 pKa = 11.84STWQRR23 pKa = 11.84NFTHH27 pKa = 5.98KK28 pKa = 9.74TSFNAGKK35 pKa = 10.27LIPFYY40 pKa = 10.25IDD42 pKa = 2.91TDD44 pKa = 4.38IIPGTTIKK52 pKa = 10.78NKK54 pKa = 7.91TSIVIRR60 pKa = 11.84MSTPIAPVMDD70 pKa = 4.2NLYY73 pKa = 10.83LDD75 pKa = 4.06TYY77 pKa = 10.16YY78 pKa = 10.85FKK80 pKa = 10.92CSKK83 pKa = 9.39FWYY86 pKa = 8.05WEE88 pKa = 3.68HH89 pKa = 6.45FRR91 pKa = 11.84AQMGEE96 pKa = 4.08NNQSAWTQTIEE107 pKa = 4.47YY108 pKa = 9.45IEE110 pKa = 4.36PQIKK114 pKa = 8.49MLSMEE119 pKa = 4.1YY120 pKa = 10.41GANDD124 pKa = 3.34VACYY128 pKa = 9.56MGMPINVANLEE139 pKa = 4.18WSKK142 pKa = 11.35LPVQAYY148 pKa = 9.67IDD150 pKa = 3.21IWNNWFRR157 pKa = 11.84DD158 pKa = 3.68EE159 pKa = 4.63NLQAPITLDD168 pKa = 3.21KK169 pKa = 11.01TDD171 pKa = 4.44ADD173 pKa = 4.01LTADD177 pKa = 3.42ATINTGFGLLPVCKK191 pKa = 9.86FHH193 pKa = 8.39DD194 pKa = 4.49YY195 pKa = 8.25FTSLLPQPQKK205 pKa = 11.38GEE207 pKa = 4.09AVTMPLGLTAPVTGTVIGNGKK228 pKa = 7.37TLGLWNGTTTAGIRR242 pKa = 11.84QGIGVSGDD250 pKa = 3.22NYY252 pKa = 11.04SNGIGIASYY261 pKa = 10.98NSSNLPYY268 pKa = 8.63NTSGVSSLNNTAGLGVTTNPDD289 pKa = 2.93KK290 pKa = 11.38SGLKK294 pKa = 9.37IASNAVADD302 pKa = 4.19LTSATAATFNALRR315 pKa = 11.84LAGATQRR322 pKa = 11.84ILEE325 pKa = 4.15KK326 pKa = 10.39DD327 pKa = 3.0ARR329 pKa = 11.84FGTRR333 pKa = 11.84YY334 pKa = 10.0PEE336 pKa = 4.04FLKK339 pKa = 10.96GQYY342 pKa = 10.34GVTAADD348 pKa = 3.67EE349 pKa = 4.4CLLIPEE355 pKa = 4.23YY356 pKa = 10.85LGGKK360 pKa = 9.42RR361 pKa = 11.84IPINIEE367 pKa = 3.84QVCQTSSTDD376 pKa = 3.15STSPLGEE383 pKa = 4.06TGAMSVTADD392 pKa = 3.13INEE395 pKa = 4.73DD396 pKa = 3.43FTKK399 pKa = 10.84SFTKK403 pKa = 10.11HH404 pKa = 6.27DD405 pKa = 3.95FLIGVCCVRR414 pKa = 11.84AEE416 pKa = 3.94HH417 pKa = 6.96TYY419 pKa = 9.94QQGIPLQFKK428 pKa = 9.92RR429 pKa = 11.84KK430 pKa = 9.03RR431 pKa = 11.84RR432 pKa = 11.84LDD434 pKa = 3.49TYY436 pKa = 10.51HH437 pKa = 7.55PSLAHH442 pKa = 6.92IGNQPVYY449 pKa = 10.03NFEE452 pKa = 4.34IFAQGNSTDD461 pKa = 4.19DD462 pKa = 3.11EE463 pKa = 4.97VMGYY467 pKa = 9.92KK468 pKa = 9.81EE469 pKa = 3.85AWQEE473 pKa = 3.68YY474 pKa = 9.17LYY476 pKa = 10.9KK477 pKa = 10.26PNRR480 pKa = 11.84ISGQMLSTYY489 pKa = 7.29TQSLDD494 pKa = 2.93VWHH497 pKa = 6.74YY498 pKa = 10.96GDD500 pKa = 5.8DD501 pKa = 4.11YY502 pKa = 12.05VNLPVLSSEE511 pKa = 4.47WIVEE515 pKa = 3.72PTEE518 pKa = 4.26YY519 pKa = 9.51IDD521 pKa = 3.24RR522 pKa = 11.84TLAVSSSVSNQFWADD537 pKa = 2.99IYY539 pKa = 10.83IEE541 pKa = 4.41QIVSAPIPINRR552 pKa = 11.84VPGLIDD558 pKa = 3.56HH559 pKa = 6.51YY560 pKa = 11.72
Molecular weight: 62.4 kDa
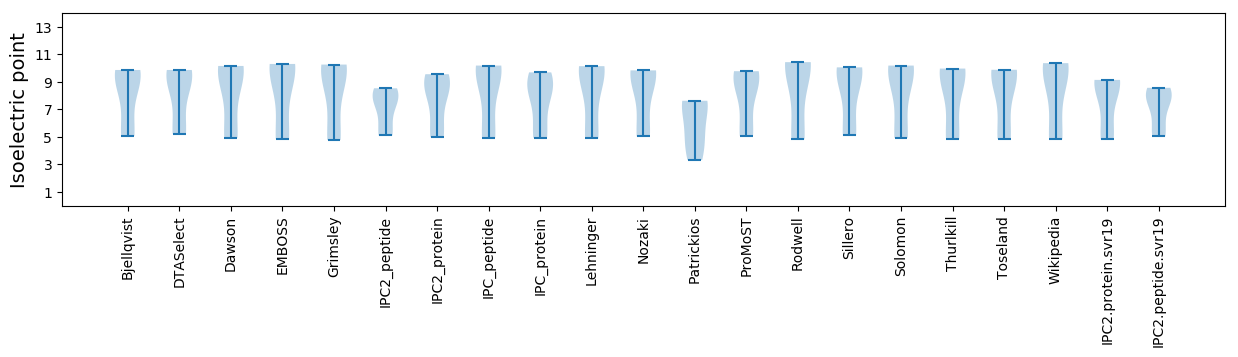
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVP3|A0A4V1FVP3_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_124 OX=2585379 PE=4 SV=1
MM1 pKa = 7.33TRR3 pKa = 11.84LSHH6 pKa = 5.15NQYY9 pKa = 9.69YY10 pKa = 10.19GEE12 pKa = 4.11RR13 pKa = 11.84KK14 pKa = 9.76NVFEE18 pKa = 4.67MLNIVYY24 pKa = 9.35RR25 pKa = 11.84KK26 pKa = 9.01PSQYY30 pKa = 10.14HH31 pKa = 5.24HH32 pKa = 7.3AGHH35 pKa = 7.07KK36 pKa = 9.21EE37 pKa = 3.87PQRR40 pKa = 11.84LSHH43 pKa = 6.24RR44 pKa = 11.84LKK46 pKa = 8.6WYY48 pKa = 9.21YY49 pKa = 11.38YY50 pKa = 10.06KK51 pKa = 10.21FTPEE55 pKa = 4.02KK56 pKa = 10.18IAIEE60 pKa = 3.91NEE62 pKa = 3.47INRR65 pKa = 11.84NQRR68 pKa = 11.84PNIQNEE74 pKa = 4.16KK75 pKa = 10.4
MM1 pKa = 7.33TRR3 pKa = 11.84LSHH6 pKa = 5.15NQYY9 pKa = 9.69YY10 pKa = 10.19GEE12 pKa = 4.11RR13 pKa = 11.84KK14 pKa = 9.76NVFEE18 pKa = 4.67MLNIVYY24 pKa = 9.35RR25 pKa = 11.84KK26 pKa = 9.01PSQYY30 pKa = 10.14HH31 pKa = 5.24HH32 pKa = 7.3AGHH35 pKa = 7.07KK36 pKa = 9.21EE37 pKa = 3.87PQRR40 pKa = 11.84LSHH43 pKa = 6.24RR44 pKa = 11.84LKK46 pKa = 8.6WYY48 pKa = 9.21YY49 pKa = 11.38YY50 pKa = 10.06KK51 pKa = 10.21FTPEE55 pKa = 4.02KK56 pKa = 10.18IAIEE60 pKa = 3.91NEE62 pKa = 3.47INRR65 pKa = 11.84NQRR68 pKa = 11.84PNIQNEE74 pKa = 4.16KK75 pKa = 10.4
Molecular weight: 9.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1502 |
75 |
560 |
250.3 |
28.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.325 ± 1.08 | 1.265 ± 0.364 |
4.594 ± 0.821 | 7.39 ± 1.182 |
3.995 ± 0.435 | 6.591 ± 1.238 |
1.598 ± 0.448 | 6.991 ± 0.746 |
9.521 ± 2.208 | 7.19 ± 0.621 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 0.385 | 7.39 ± 0.512 |
3.662 ± 0.743 | 3.995 ± 0.245 |
4.194 ± 0.565 | 5.992 ± 1.357 |
7.523 ± 0.733 | 3.129 ± 0.836 |
1.265 ± 0.376 | 4.794 ± 0.442 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
