
Botryotinia fuckeliana totivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Totivirus; unclassified Totivirus
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
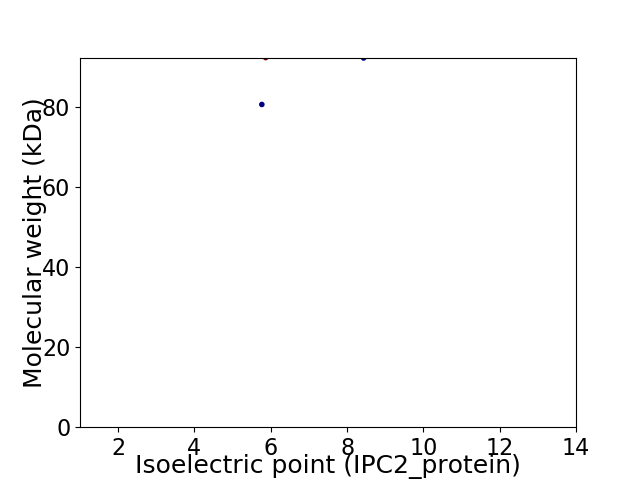
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
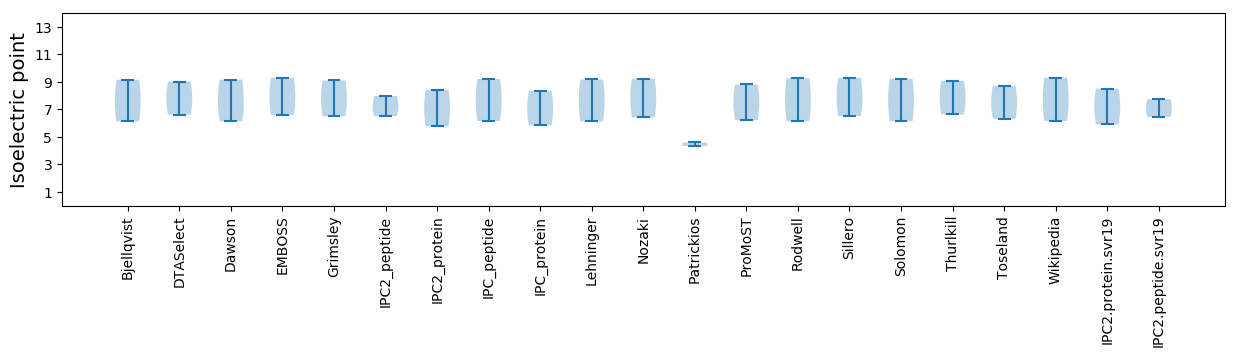
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4FRF6|A4FRF6_9VIRU RNA-directed RNA polymerase OS=Botryotinia fuckeliana totivirus 1 OX=425009 GN=RdRp PE=3 SV=1
MM1 pKa = 7.71SSQLNPTFLSGVLANARR18 pKa = 11.84GGRR21 pKa = 11.84ITTKK25 pKa = 9.73DD26 pKa = 2.57TYY28 pKa = 10.88RR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 8.79KK32 pKa = 10.18TIVRR36 pKa = 11.84TSATIAGVLDD46 pKa = 3.57TRR48 pKa = 11.84LTGIFYY54 pKa = 10.12EE55 pKa = 4.25IGRR58 pKa = 11.84AVDD61 pKa = 3.49TKK63 pKa = 11.14GKK65 pKa = 9.74LLKK68 pKa = 10.08SHH70 pKa = 6.98KK71 pKa = 10.23EE72 pKa = 3.92DD73 pKa = 3.08ITLVEE78 pKa = 4.28ASYY81 pKa = 8.46PTNSVLAEE89 pKa = 4.3DD90 pKa = 5.33FIGMAKK96 pKa = 10.32KK97 pKa = 8.94YY98 pKa = 8.7TNFSASFEE106 pKa = 4.23YY107 pKa = 10.72SSLAGVVEE115 pKa = 4.18RR116 pKa = 11.84LGKK119 pKa = 10.5GLAACSLFDD128 pKa = 4.53SVTSTDD134 pKa = 3.34LRR136 pKa = 11.84GGAVLAVNALGTYY149 pKa = 9.7DD150 pKa = 4.08GPVNSLTDD158 pKa = 3.24AVYY161 pKa = 10.19IPRR164 pKa = 11.84LVNTTLTGDD173 pKa = 3.89VFAVLCNAVAGEE185 pKa = 4.54GSQVVSDD192 pKa = 4.33VVEE195 pKa = 4.18LDD197 pKa = 3.1ANTRR201 pKa = 11.84QPIIALVDD209 pKa = 3.87SVGLPGAIVDD219 pKa = 3.98ALRR222 pKa = 11.84VLGSNMIASDD232 pKa = 3.62QGPLFSLALARR243 pKa = 11.84GLHH246 pKa = 6.33RR247 pKa = 11.84GLSLVGHH254 pKa = 5.99TDD256 pKa = 2.87EE257 pKa = 6.06GSITRR262 pKa = 11.84DD263 pKa = 3.25LLRR266 pKa = 11.84CGGFAPPFGGIHH278 pKa = 6.3YY279 pKa = 10.33GLEE282 pKa = 4.37PYY284 pKa = 10.51VGLPALQTNLDD295 pKa = 4.12KK296 pKa = 11.28GVSAYY301 pKa = 10.79VDD303 pKa = 4.18AIVIATGAAVAHH315 pKa = 7.07ADD317 pKa = 3.17PGTRR321 pKa = 11.84FDD323 pKa = 3.92GHH325 pKa = 6.75WFPTFISGTASGEE338 pKa = 3.81PLLRR342 pKa = 11.84PGEE345 pKa = 4.33HH346 pKa = 5.24TTGTTAMADD355 pKa = 4.17RR356 pKa = 11.84IRR358 pKa = 11.84ARR360 pKa = 11.84LLADD364 pKa = 3.62QSPFWEE370 pKa = 5.9GYY372 pKa = 7.77TRR374 pKa = 11.84ALSIIFGAEE383 pKa = 3.63GDD385 pKa = 3.69TSLSVRR391 pKa = 11.84FACAASTALDD401 pKa = 3.28VDD403 pKa = 3.75NRR405 pKa = 11.84HH406 pKa = 6.28LKK408 pKa = 9.19HH409 pKa = 6.66ASVSPWFWVEE419 pKa = 3.74PTSILPPNLLGSEE432 pKa = 4.39AEE434 pKa = 4.41LEE436 pKa = 4.67GCASLAHH443 pKa = 7.03KK444 pKa = 9.32DD445 pKa = 3.71TVVSKK450 pKa = 9.35PAWDD454 pKa = 3.89TITRR458 pKa = 11.84VGSSDD463 pKa = 3.41VSFSAYY469 pKa = 9.45RR470 pKa = 11.84ARR472 pKa = 11.84FKK474 pKa = 10.54FARR477 pKa = 11.84QTWFFNHH484 pKa = 6.57WLNHH488 pKa = 4.87PMNGLGAMKK497 pKa = 10.08VRR499 pKa = 11.84QLDD502 pKa = 3.65PNGIIHH508 pKa = 7.11PGPCTAHH515 pKa = 5.62PQVRR519 pKa = 11.84DD520 pKa = 3.33RR521 pKa = 11.84VEE523 pKa = 4.5ADD525 pKa = 4.14LPFTDD530 pKa = 4.37YY531 pKa = 11.4LWIRR535 pKa = 11.84GQSPFPAPGEE545 pKa = 4.17LINLGLGVGFLVRR558 pKa = 11.84HH559 pKa = 5.11MTLDD563 pKa = 3.34AEE565 pKa = 4.84GIPNEE570 pKa = 4.02EE571 pKa = 4.32HH572 pKa = 6.77LPTSRR577 pKa = 11.84EE578 pKa = 3.76IQDD581 pKa = 3.76CVVTIDD587 pKa = 3.11VGRR590 pKa = 11.84PIGLASGKK598 pKa = 8.58TNAADD603 pKa = 3.78SQAKK607 pKa = 9.35RR608 pKa = 11.84ARR610 pKa = 11.84TRR612 pKa = 11.84ANNEE616 pKa = 3.27LAAARR621 pKa = 11.84TRR623 pKa = 11.84ARR625 pKa = 11.84VFGTPDD631 pKa = 3.13VGEE634 pKa = 4.44MPTLTTAPGYY644 pKa = 7.16ATRR647 pKa = 11.84WSSEE651 pKa = 3.57QGIDD655 pKa = 3.51STSGGVVSRR664 pKa = 11.84GRR666 pKa = 11.84HH667 pKa = 4.21DD668 pKa = 3.44HH669 pKa = 6.6AAPGHH674 pKa = 6.12ASGGDD679 pKa = 3.36RR680 pKa = 11.84EE681 pKa = 4.53PTGKK685 pKa = 10.2QNIAVPQHH693 pKa = 5.17QPLRR697 pKa = 11.84YY698 pKa = 8.31PALPRR703 pKa = 11.84QGAGLGGGANPIPPPPAPPAGGPDD727 pKa = 4.87DD728 pKa = 5.66GPQSDD733 pKa = 5.39DD734 pKa = 4.61DD735 pKa = 4.28NTPSAPTAPLGLSTTPPRR753 pKa = 11.84SPAAPGNGPDD763 pKa = 3.96TII765 pKa = 4.52
MM1 pKa = 7.71SSQLNPTFLSGVLANARR18 pKa = 11.84GGRR21 pKa = 11.84ITTKK25 pKa = 9.73DD26 pKa = 2.57TYY28 pKa = 10.88RR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 8.79KK32 pKa = 10.18TIVRR36 pKa = 11.84TSATIAGVLDD46 pKa = 3.57TRR48 pKa = 11.84LTGIFYY54 pKa = 10.12EE55 pKa = 4.25IGRR58 pKa = 11.84AVDD61 pKa = 3.49TKK63 pKa = 11.14GKK65 pKa = 9.74LLKK68 pKa = 10.08SHH70 pKa = 6.98KK71 pKa = 10.23EE72 pKa = 3.92DD73 pKa = 3.08ITLVEE78 pKa = 4.28ASYY81 pKa = 8.46PTNSVLAEE89 pKa = 4.3DD90 pKa = 5.33FIGMAKK96 pKa = 10.32KK97 pKa = 8.94YY98 pKa = 8.7TNFSASFEE106 pKa = 4.23YY107 pKa = 10.72SSLAGVVEE115 pKa = 4.18RR116 pKa = 11.84LGKK119 pKa = 10.5GLAACSLFDD128 pKa = 4.53SVTSTDD134 pKa = 3.34LRR136 pKa = 11.84GGAVLAVNALGTYY149 pKa = 9.7DD150 pKa = 4.08GPVNSLTDD158 pKa = 3.24AVYY161 pKa = 10.19IPRR164 pKa = 11.84LVNTTLTGDD173 pKa = 3.89VFAVLCNAVAGEE185 pKa = 4.54GSQVVSDD192 pKa = 4.33VVEE195 pKa = 4.18LDD197 pKa = 3.1ANTRR201 pKa = 11.84QPIIALVDD209 pKa = 3.87SVGLPGAIVDD219 pKa = 3.98ALRR222 pKa = 11.84VLGSNMIASDD232 pKa = 3.62QGPLFSLALARR243 pKa = 11.84GLHH246 pKa = 6.33RR247 pKa = 11.84GLSLVGHH254 pKa = 5.99TDD256 pKa = 2.87EE257 pKa = 6.06GSITRR262 pKa = 11.84DD263 pKa = 3.25LLRR266 pKa = 11.84CGGFAPPFGGIHH278 pKa = 6.3YY279 pKa = 10.33GLEE282 pKa = 4.37PYY284 pKa = 10.51VGLPALQTNLDD295 pKa = 4.12KK296 pKa = 11.28GVSAYY301 pKa = 10.79VDD303 pKa = 4.18AIVIATGAAVAHH315 pKa = 7.07ADD317 pKa = 3.17PGTRR321 pKa = 11.84FDD323 pKa = 3.92GHH325 pKa = 6.75WFPTFISGTASGEE338 pKa = 3.81PLLRR342 pKa = 11.84PGEE345 pKa = 4.33HH346 pKa = 5.24TTGTTAMADD355 pKa = 4.17RR356 pKa = 11.84IRR358 pKa = 11.84ARR360 pKa = 11.84LLADD364 pKa = 3.62QSPFWEE370 pKa = 5.9GYY372 pKa = 7.77TRR374 pKa = 11.84ALSIIFGAEE383 pKa = 3.63GDD385 pKa = 3.69TSLSVRR391 pKa = 11.84FACAASTALDD401 pKa = 3.28VDD403 pKa = 3.75NRR405 pKa = 11.84HH406 pKa = 6.28LKK408 pKa = 9.19HH409 pKa = 6.66ASVSPWFWVEE419 pKa = 3.74PTSILPPNLLGSEE432 pKa = 4.39AEE434 pKa = 4.41LEE436 pKa = 4.67GCASLAHH443 pKa = 7.03KK444 pKa = 9.32DD445 pKa = 3.71TVVSKK450 pKa = 9.35PAWDD454 pKa = 3.89TITRR458 pKa = 11.84VGSSDD463 pKa = 3.41VSFSAYY469 pKa = 9.45RR470 pKa = 11.84ARR472 pKa = 11.84FKK474 pKa = 10.54FARR477 pKa = 11.84QTWFFNHH484 pKa = 6.57WLNHH488 pKa = 4.87PMNGLGAMKK497 pKa = 10.08VRR499 pKa = 11.84QLDD502 pKa = 3.65PNGIIHH508 pKa = 7.11PGPCTAHH515 pKa = 5.62PQVRR519 pKa = 11.84DD520 pKa = 3.33RR521 pKa = 11.84VEE523 pKa = 4.5ADD525 pKa = 4.14LPFTDD530 pKa = 4.37YY531 pKa = 11.4LWIRR535 pKa = 11.84GQSPFPAPGEE545 pKa = 4.17LINLGLGVGFLVRR558 pKa = 11.84HH559 pKa = 5.11MTLDD563 pKa = 3.34AEE565 pKa = 4.84GIPNEE570 pKa = 4.02EE571 pKa = 4.32HH572 pKa = 6.77LPTSRR577 pKa = 11.84EE578 pKa = 3.76IQDD581 pKa = 3.76CVVTIDD587 pKa = 3.11VGRR590 pKa = 11.84PIGLASGKK598 pKa = 8.58TNAADD603 pKa = 3.78SQAKK607 pKa = 9.35RR608 pKa = 11.84ARR610 pKa = 11.84TRR612 pKa = 11.84ANNEE616 pKa = 3.27LAAARR621 pKa = 11.84TRR623 pKa = 11.84ARR625 pKa = 11.84VFGTPDD631 pKa = 3.13VGEE634 pKa = 4.44MPTLTTAPGYY644 pKa = 7.16ATRR647 pKa = 11.84WSSEE651 pKa = 3.57QGIDD655 pKa = 3.51STSGGVVSRR664 pKa = 11.84GRR666 pKa = 11.84HH667 pKa = 4.21DD668 pKa = 3.44HH669 pKa = 6.6AAPGHH674 pKa = 6.12ASGGDD679 pKa = 3.36RR680 pKa = 11.84EE681 pKa = 4.53PTGKK685 pKa = 10.2QNIAVPQHH693 pKa = 5.17QPLRR697 pKa = 11.84YY698 pKa = 8.31PALPRR703 pKa = 11.84QGAGLGGGANPIPPPPAPPAGGPDD727 pKa = 4.87DD728 pKa = 5.66GPQSDD733 pKa = 5.39DD734 pKa = 4.61DD735 pKa = 4.28NTPSAPTAPLGLSTTPPRR753 pKa = 11.84SPAAPGNGPDD763 pKa = 3.96TII765 pKa = 4.52
Molecular weight: 80.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4FRF6|A4FRF6_9VIRU RNA-directed RNA polymerase OS=Botryotinia fuckeliana totivirus 1 OX=425009 GN=RdRp PE=3 SV=1
MM1 pKa = 7.3SAADD5 pKa = 3.72RR6 pKa = 11.84ASAYY10 pKa = 10.17GRR12 pKa = 11.84LGSYY16 pKa = 10.28LKK18 pKa = 10.57EE19 pKa = 4.34LLDD22 pKa = 3.97DD23 pKa = 4.22HH24 pKa = 7.14PFVQGLFSTTNFTQSLIRR42 pKa = 11.84LQGSSFTMRR51 pKa = 11.84SAHH54 pKa = 6.73PLLPHH59 pKa = 6.88AASLLLLDD67 pKa = 6.0FPMQTDD73 pKa = 3.76TSVSDD78 pKa = 4.13VISLARR84 pKa = 11.84NAYY87 pKa = 9.56DD88 pKa = 4.94LSDD91 pKa = 3.88LGEE94 pKa = 4.51DD95 pKa = 3.41VLINHH100 pKa = 6.92TLLPPGSPILSWRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84GYY117 pKa = 9.79HH118 pKa = 6.15RR119 pKa = 11.84KK120 pKa = 9.72LGNAIITNKK129 pKa = 8.69EE130 pKa = 3.79LRR132 pKa = 11.84DD133 pKa = 3.5SLFPKK138 pKa = 9.9KK139 pKa = 9.99KK140 pKa = 9.98HH141 pKa = 5.31NAAGVKK147 pKa = 10.13VNLTIGRR154 pKa = 11.84LAVAWARR161 pKa = 11.84VFGPGSLGRR170 pKa = 11.84HH171 pKa = 5.57LAAIAGRR178 pKa = 11.84VTNDD182 pKa = 3.25QACSALLYY190 pKa = 10.63CLTLRR195 pKa = 11.84EE196 pKa = 4.66HH197 pKa = 6.66IGSFGISIAQAAVTQPANAKK217 pKa = 9.8GLSNALKK224 pKa = 10.61ALGANSSLPGSLLVEE239 pKa = 4.38AATLQGRR246 pKa = 11.84FTNDD250 pKa = 1.91VDD252 pKa = 3.64MTNEE256 pKa = 3.4IRR258 pKa = 11.84SRR260 pKa = 11.84TIQSLVDD267 pKa = 3.43EE268 pKa = 4.43QVIDD272 pKa = 3.71RR273 pKa = 11.84PEE275 pKa = 3.76EE276 pKa = 3.76LRR278 pKa = 11.84PHH280 pKa = 6.48IKK282 pKa = 10.54ALLEE286 pKa = 3.76MSLPGDD292 pKa = 3.78CALPDD297 pKa = 3.53MDD299 pKa = 4.75EE300 pKa = 4.09WWSSRR305 pKa = 11.84WLWCVNGSEE314 pKa = 4.45TKK316 pKa = 10.65KK317 pKa = 10.67SDD319 pKa = 3.16EE320 pKa = 4.05ALGLRR325 pKa = 11.84GGSGRR330 pKa = 11.84RR331 pKa = 11.84YY332 pKa = 9.24RR333 pKa = 11.84RR334 pKa = 11.84MAAEE338 pKa = 4.31EE339 pKa = 4.16VHH341 pKa = 6.71SNPVPDD347 pKa = 3.61WDD349 pKa = 3.84GTTSVSASIKK359 pKa = 10.73LEE361 pKa = 4.13AGKK364 pKa = 10.4DD365 pKa = 3.39RR366 pKa = 11.84AIFACDD372 pKa = 3.05TRR374 pKa = 11.84SYY376 pKa = 11.08FAFSWILNDD385 pKa = 4.35VQKK388 pKa = 10.49KK389 pKa = 6.96WKK391 pKa = 10.19GEE393 pKa = 4.16RR394 pKa = 11.84IPLDD398 pKa = 3.79PGKK401 pKa = 10.75GGLYY405 pKa = 10.22GISRR409 pKa = 11.84RR410 pKa = 11.84IRR412 pKa = 11.84NSQRR416 pKa = 11.84GGGVNLMLDD425 pKa = 3.48YY426 pKa = 11.6DD427 pKa = 4.41NFNSHH432 pKa = 6.79HH433 pKa = 6.59SNSVQSMIFEE443 pKa = 4.38VLCDD447 pKa = 4.44KK448 pKa = 11.25YY449 pKa = 11.22NAPQWYY455 pKa = 9.66KK456 pKa = 10.27EE457 pKa = 3.79VLMEE461 pKa = 4.46SFNRR465 pKa = 11.84MYY467 pKa = 10.39IFKK470 pKa = 10.56GGVRR474 pKa = 11.84HH475 pKa = 6.73RR476 pKa = 11.84MLGTLASGHH485 pKa = 6.67RR486 pKa = 11.84GTSFINSLLNAAYY499 pKa = 9.36IRR501 pKa = 11.84CALGAAKK508 pKa = 9.93FDD510 pKa = 3.99TMLSLHH516 pKa = 6.87AGDD519 pKa = 4.95DD520 pKa = 3.76VYY522 pKa = 11.3IRR524 pKa = 11.84ANTLSEE530 pKa = 4.05AADD533 pKa = 3.34ILKK536 pKa = 9.36KK537 pKa = 10.73CKK539 pKa = 9.85MFGCRR544 pKa = 11.84MNPTKK549 pKa = 10.58QSIGFRR555 pKa = 11.84NAEE558 pKa = 4.11FLRR561 pKa = 11.84LGINSRR567 pKa = 11.84YY568 pKa = 9.44AVGYY572 pKa = 6.92VARR575 pKa = 11.84TISTLVAGNWSNLDD589 pKa = 3.19PMEE592 pKa = 4.56PLEE595 pKa = 4.49SLTSVISSVRR605 pKa = 11.84SVINRR610 pKa = 11.84GAPTIIADD618 pKa = 4.29VIAYY622 pKa = 8.64AHH624 pKa = 6.09STLHH628 pKa = 6.66PYY630 pKa = 8.78PLKK633 pKa = 10.35ILKK636 pKa = 9.48PLLRR640 pKa = 11.84GEE642 pKa = 4.1AAIAGAPIFNLHH654 pKa = 5.14GKK656 pKa = 7.79MRR658 pKa = 11.84TYY660 pKa = 10.44EE661 pKa = 4.11AVVSKK666 pKa = 10.25PDD668 pKa = 3.4NSGNPPPPEE677 pKa = 4.23WGSHH681 pKa = 4.08ATLSYY686 pKa = 10.97LEE688 pKa = 4.21YY689 pKa = 10.37HH690 pKa = 6.2VAPIEE695 pKa = 3.97ASAISRR701 pKa = 11.84ARR703 pKa = 11.84VDD705 pKa = 3.55LTSIMQEE712 pKa = 3.32SSYY715 pKa = 11.19AKK717 pKa = 9.55QVRR720 pKa = 11.84SDD722 pKa = 3.86LDD724 pKa = 3.53DD725 pKa = 4.13ASRR728 pKa = 11.84RR729 pKa = 11.84TTVKK733 pKa = 10.7LIPRR737 pKa = 11.84TPEE740 pKa = 3.4EE741 pKa = 3.58ARR743 pKa = 11.84GYY745 pKa = 10.26IDD747 pKa = 5.58ASTLSTRR754 pKa = 11.84DD755 pKa = 3.14AKK757 pKa = 10.77RR758 pKa = 11.84GILEE762 pKa = 4.24SYY764 pKa = 10.04PLIRR768 pKa = 11.84LLEE771 pKa = 4.17NRR773 pKa = 11.84LSDD776 pKa = 3.44EE777 pKa = 4.49DD778 pKa = 3.97LRR780 pKa = 11.84EE781 pKa = 3.96LALLLGVSCSTKK793 pKa = 9.4EE794 pKa = 4.34ARR796 pKa = 11.84VCCFGSEE803 pKa = 4.27SLTHH807 pKa = 6.59NIIGYY812 pKa = 9.47LPYY815 pKa = 10.56SDD817 pKa = 4.55AASMSKK823 pKa = 7.23TTNSRR828 pKa = 11.84NIYY831 pKa = 5.19TTYY834 pKa = 10.46HH835 pKa = 4.55VRR837 pKa = 11.84AA838 pKa = 4.25
MM1 pKa = 7.3SAADD5 pKa = 3.72RR6 pKa = 11.84ASAYY10 pKa = 10.17GRR12 pKa = 11.84LGSYY16 pKa = 10.28LKK18 pKa = 10.57EE19 pKa = 4.34LLDD22 pKa = 3.97DD23 pKa = 4.22HH24 pKa = 7.14PFVQGLFSTTNFTQSLIRR42 pKa = 11.84LQGSSFTMRR51 pKa = 11.84SAHH54 pKa = 6.73PLLPHH59 pKa = 6.88AASLLLLDD67 pKa = 6.0FPMQTDD73 pKa = 3.76TSVSDD78 pKa = 4.13VISLARR84 pKa = 11.84NAYY87 pKa = 9.56DD88 pKa = 4.94LSDD91 pKa = 3.88LGEE94 pKa = 4.51DD95 pKa = 3.41VLINHH100 pKa = 6.92TLLPPGSPILSWRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84GYY117 pKa = 9.79HH118 pKa = 6.15RR119 pKa = 11.84KK120 pKa = 9.72LGNAIITNKK129 pKa = 8.69EE130 pKa = 3.79LRR132 pKa = 11.84DD133 pKa = 3.5SLFPKK138 pKa = 9.9KK139 pKa = 9.99KK140 pKa = 9.98HH141 pKa = 5.31NAAGVKK147 pKa = 10.13VNLTIGRR154 pKa = 11.84LAVAWARR161 pKa = 11.84VFGPGSLGRR170 pKa = 11.84HH171 pKa = 5.57LAAIAGRR178 pKa = 11.84VTNDD182 pKa = 3.25QACSALLYY190 pKa = 10.63CLTLRR195 pKa = 11.84EE196 pKa = 4.66HH197 pKa = 6.66IGSFGISIAQAAVTQPANAKK217 pKa = 9.8GLSNALKK224 pKa = 10.61ALGANSSLPGSLLVEE239 pKa = 4.38AATLQGRR246 pKa = 11.84FTNDD250 pKa = 1.91VDD252 pKa = 3.64MTNEE256 pKa = 3.4IRR258 pKa = 11.84SRR260 pKa = 11.84TIQSLVDD267 pKa = 3.43EE268 pKa = 4.43QVIDD272 pKa = 3.71RR273 pKa = 11.84PEE275 pKa = 3.76EE276 pKa = 3.76LRR278 pKa = 11.84PHH280 pKa = 6.48IKK282 pKa = 10.54ALLEE286 pKa = 3.76MSLPGDD292 pKa = 3.78CALPDD297 pKa = 3.53MDD299 pKa = 4.75EE300 pKa = 4.09WWSSRR305 pKa = 11.84WLWCVNGSEE314 pKa = 4.45TKK316 pKa = 10.65KK317 pKa = 10.67SDD319 pKa = 3.16EE320 pKa = 4.05ALGLRR325 pKa = 11.84GGSGRR330 pKa = 11.84RR331 pKa = 11.84YY332 pKa = 9.24RR333 pKa = 11.84RR334 pKa = 11.84MAAEE338 pKa = 4.31EE339 pKa = 4.16VHH341 pKa = 6.71SNPVPDD347 pKa = 3.61WDD349 pKa = 3.84GTTSVSASIKK359 pKa = 10.73LEE361 pKa = 4.13AGKK364 pKa = 10.4DD365 pKa = 3.39RR366 pKa = 11.84AIFACDD372 pKa = 3.05TRR374 pKa = 11.84SYY376 pKa = 11.08FAFSWILNDD385 pKa = 4.35VQKK388 pKa = 10.49KK389 pKa = 6.96WKK391 pKa = 10.19GEE393 pKa = 4.16RR394 pKa = 11.84IPLDD398 pKa = 3.79PGKK401 pKa = 10.75GGLYY405 pKa = 10.22GISRR409 pKa = 11.84RR410 pKa = 11.84IRR412 pKa = 11.84NSQRR416 pKa = 11.84GGGVNLMLDD425 pKa = 3.48YY426 pKa = 11.6DD427 pKa = 4.41NFNSHH432 pKa = 6.79HH433 pKa = 6.59SNSVQSMIFEE443 pKa = 4.38VLCDD447 pKa = 4.44KK448 pKa = 11.25YY449 pKa = 11.22NAPQWYY455 pKa = 9.66KK456 pKa = 10.27EE457 pKa = 3.79VLMEE461 pKa = 4.46SFNRR465 pKa = 11.84MYY467 pKa = 10.39IFKK470 pKa = 10.56GGVRR474 pKa = 11.84HH475 pKa = 6.73RR476 pKa = 11.84MLGTLASGHH485 pKa = 6.67RR486 pKa = 11.84GTSFINSLLNAAYY499 pKa = 9.36IRR501 pKa = 11.84CALGAAKK508 pKa = 9.93FDD510 pKa = 3.99TMLSLHH516 pKa = 6.87AGDD519 pKa = 4.95DD520 pKa = 3.76VYY522 pKa = 11.3IRR524 pKa = 11.84ANTLSEE530 pKa = 4.05AADD533 pKa = 3.34ILKK536 pKa = 9.36KK537 pKa = 10.73CKK539 pKa = 9.85MFGCRR544 pKa = 11.84MNPTKK549 pKa = 10.58QSIGFRR555 pKa = 11.84NAEE558 pKa = 4.11FLRR561 pKa = 11.84LGINSRR567 pKa = 11.84YY568 pKa = 9.44AVGYY572 pKa = 6.92VARR575 pKa = 11.84TISTLVAGNWSNLDD589 pKa = 3.19PMEE592 pKa = 4.56PLEE595 pKa = 4.49SLTSVISSVRR605 pKa = 11.84SVINRR610 pKa = 11.84GAPTIIADD618 pKa = 4.29VIAYY622 pKa = 8.64AHH624 pKa = 6.09STLHH628 pKa = 6.66PYY630 pKa = 8.78PLKK633 pKa = 10.35ILKK636 pKa = 9.48PLLRR640 pKa = 11.84GEE642 pKa = 4.1AAIAGAPIFNLHH654 pKa = 5.14GKK656 pKa = 7.79MRR658 pKa = 11.84TYY660 pKa = 10.44EE661 pKa = 4.11AVVSKK666 pKa = 10.25PDD668 pKa = 3.4NSGNPPPPEE677 pKa = 4.23WGSHH681 pKa = 4.08ATLSYY686 pKa = 10.97LEE688 pKa = 4.21YY689 pKa = 10.37HH690 pKa = 6.2VAPIEE695 pKa = 3.97ASAISRR701 pKa = 11.84ARR703 pKa = 11.84VDD705 pKa = 3.55LTSIMQEE712 pKa = 3.32SSYY715 pKa = 11.19AKK717 pKa = 9.55QVRR720 pKa = 11.84SDD722 pKa = 3.86LDD724 pKa = 3.53DD725 pKa = 4.13ASRR728 pKa = 11.84RR729 pKa = 11.84TTVKK733 pKa = 10.7LIPRR737 pKa = 11.84TPEE740 pKa = 3.4EE741 pKa = 3.58ARR743 pKa = 11.84GYY745 pKa = 10.26IDD747 pKa = 5.58ASTLSTRR754 pKa = 11.84DD755 pKa = 3.14AKK757 pKa = 10.77RR758 pKa = 11.84GILEE762 pKa = 4.24SYY764 pKa = 10.04PLIRR768 pKa = 11.84LLEE771 pKa = 4.17NRR773 pKa = 11.84LSDD776 pKa = 3.44EE777 pKa = 4.49DD778 pKa = 3.97LRR780 pKa = 11.84EE781 pKa = 3.96LALLLGVSCSTKK793 pKa = 9.4EE794 pKa = 4.34ARR796 pKa = 11.84VCCFGSEE803 pKa = 4.27SLTHH807 pKa = 6.59NIIGYY812 pKa = 9.47LPYY815 pKa = 10.56SDD817 pKa = 4.55AASMSKK823 pKa = 7.23TTNSRR828 pKa = 11.84NIYY831 pKa = 5.19TTYY834 pKa = 10.46HH835 pKa = 4.55VRR837 pKa = 11.84AA838 pKa = 4.25
Molecular weight: 92.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1603 |
765 |
838 |
801.5 |
86.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.168 ± 0.527 | 1.185 ± 0.175 |
5.677 ± 0.303 | 4.117 ± 0.297 |
3.057 ± 0.222 | 8.734 ± 1.204 |
2.62 ± 0.004 | 5.115 ± 0.436 |
3.306 ± 0.619 | 10.168 ± 0.746 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.684 ± 0.415 | 4.055 ± 0.426 |
6.051 ± 1.079 | 2.183 ± 0.11 |
6.925 ± 0.422 | 8.609 ± 0.922 |
6.488 ± 0.88 | 5.802 ± 0.647 |
1.31 ± 0.087 | 2.745 ± 0.424 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
