
Polyporus arcularius HHB13444
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporaceae; Polyporus; Polyporus arcularius
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
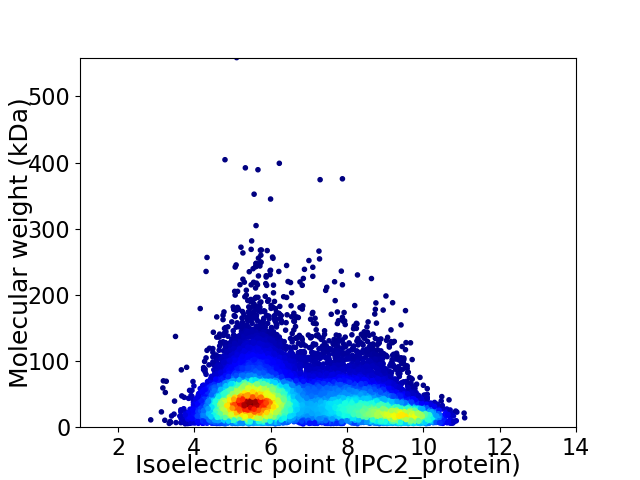
Virtual 2D-PAGE plot for 17473 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C3PB19|A0A5C3PB19_9APHY Peroxidase OS=Polyporus arcularius HHB13444 OX=1314778 GN=K466DRAFT_549730 PE=3 SV=1
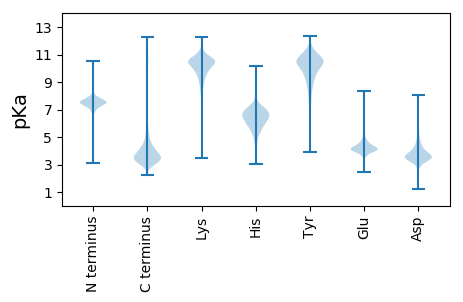
MM1 pKa = 7.37LTPHH5 pKa = 7.56RR6 pKa = 11.84WPSSCVSGHH15 pKa = 4.72VSAVRR20 pKa = 11.84GSLGIYY26 pKa = 9.66NPLRR30 pKa = 11.84EE31 pKa = 4.05RR32 pKa = 11.84FISNHH37 pKa = 5.6RR38 pKa = 11.84ASGSSISTSSSQASNQTLSVSSITSSANAPDD69 pKa = 4.46APSSTISIVSSTDD82 pKa = 2.99LATSVVATDD91 pKa = 3.68SADD94 pKa = 3.58PTDD97 pKa = 3.79SSVITISTATDD108 pKa = 4.16FFNGTATEE116 pKa = 4.4SSSPTATDD124 pKa = 3.23TTTDD128 pKa = 2.98ASATDD133 pKa = 3.97VSALPTSTDD142 pKa = 3.41DD143 pKa = 5.03ASATDD148 pKa = 3.57SSSPSTATDD157 pKa = 3.37SDD159 pKa = 4.07PGTSVSTDD167 pKa = 2.81AAAPTGTGISFDD179 pKa = 4.51LPTNIPPACLALASGTVISTPSLTSAAASATDD211 pKa = 3.8SSAADD216 pKa = 3.71PSSTALSATDD226 pKa = 3.86ASATEE231 pKa = 4.52PATADD236 pKa = 3.52PSATDD241 pKa = 3.63SLNSTSIVVTSSIPSSSISSSAAAVNVTATAIATVTDD278 pKa = 4.05VSSSAATASPTDD290 pKa = 3.28GSTNSTISRR299 pKa = 11.84RR300 pKa = 11.84IAQTDD305 pKa = 4.0LPDD308 pKa = 4.22VAQSWQDD315 pKa = 3.23LCLVSGGDD323 pKa = 3.18IFTHH327 pKa = 6.05EE328 pKa = 4.41PCVTLAGVDD337 pKa = 4.45GINALLGDD345 pKa = 5.2ADD347 pKa = 4.17PCAQQDD353 pKa = 3.75NADD356 pKa = 3.63AMIDD360 pKa = 3.8FAKK363 pKa = 10.85SPGVTNADD371 pKa = 3.34ALIANAIAYY380 pKa = 7.5RR381 pKa = 11.84QHH383 pKa = 6.39PRR385 pKa = 11.84NAINVNGVVPATPYY399 pKa = 9.81CQRR402 pKa = 11.84APRR405 pKa = 11.84NAEE408 pKa = 3.48LQGIVNTQLDD418 pKa = 4.11GVNAGIYY425 pKa = 10.49GSVNLGLYY433 pKa = 10.61AFGAAGTCPFGQSPDD448 pKa = 3.34VSTCICSS455 pKa = 3.13
MM1 pKa = 7.37LTPHH5 pKa = 7.56RR6 pKa = 11.84WPSSCVSGHH15 pKa = 4.72VSAVRR20 pKa = 11.84GSLGIYY26 pKa = 9.66NPLRR30 pKa = 11.84EE31 pKa = 4.05RR32 pKa = 11.84FISNHH37 pKa = 5.6RR38 pKa = 11.84ASGSSISTSSSQASNQTLSVSSITSSANAPDD69 pKa = 4.46APSSTISIVSSTDD82 pKa = 2.99LATSVVATDD91 pKa = 3.68SADD94 pKa = 3.58PTDD97 pKa = 3.79SSVITISTATDD108 pKa = 4.16FFNGTATEE116 pKa = 4.4SSSPTATDD124 pKa = 3.23TTTDD128 pKa = 2.98ASATDD133 pKa = 3.97VSALPTSTDD142 pKa = 3.41DD143 pKa = 5.03ASATDD148 pKa = 3.57SSSPSTATDD157 pKa = 3.37SDD159 pKa = 4.07PGTSVSTDD167 pKa = 2.81AAAPTGTGISFDD179 pKa = 4.51LPTNIPPACLALASGTVISTPSLTSAAASATDD211 pKa = 3.8SSAADD216 pKa = 3.71PSSTALSATDD226 pKa = 3.86ASATEE231 pKa = 4.52PATADD236 pKa = 3.52PSATDD241 pKa = 3.63SLNSTSIVVTSSIPSSSISSSAAAVNVTATAIATVTDD278 pKa = 4.05VSSSAATASPTDD290 pKa = 3.28GSTNSTISRR299 pKa = 11.84RR300 pKa = 11.84IAQTDD305 pKa = 4.0LPDD308 pKa = 4.22VAQSWQDD315 pKa = 3.23LCLVSGGDD323 pKa = 3.18IFTHH327 pKa = 6.05EE328 pKa = 4.41PCVTLAGVDD337 pKa = 4.45GINALLGDD345 pKa = 5.2ADD347 pKa = 4.17PCAQQDD353 pKa = 3.75NADD356 pKa = 3.63AMIDD360 pKa = 3.8FAKK363 pKa = 10.85SPGVTNADD371 pKa = 3.34ALIANAIAYY380 pKa = 7.5RR381 pKa = 11.84QHH383 pKa = 6.39PRR385 pKa = 11.84NAINVNGVVPATPYY399 pKa = 9.81CQRR402 pKa = 11.84APRR405 pKa = 11.84NAEE408 pKa = 3.48LQGIVNTQLDD418 pKa = 4.11GVNAGIYY425 pKa = 10.49GSVNLGLYY433 pKa = 10.61AFGAAGTCPFGQSPDD448 pKa = 3.34VSTCICSS455 pKa = 3.13
Molecular weight: 45.12 kDa
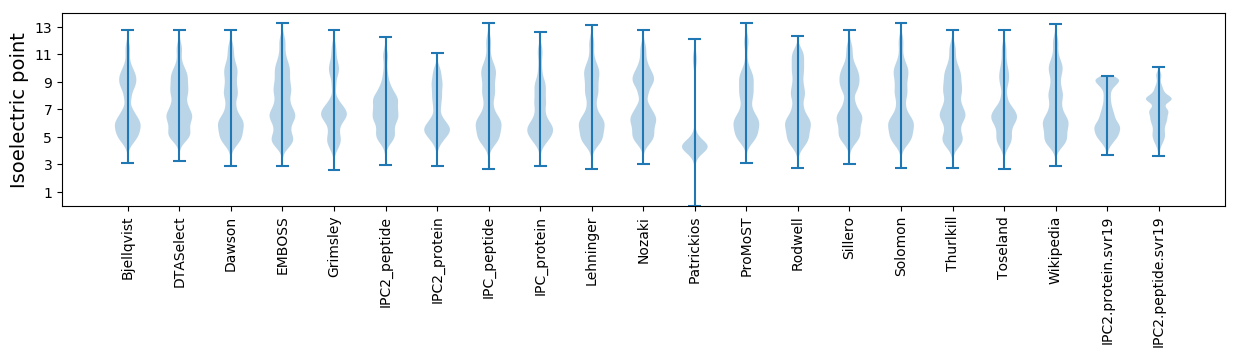
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C3PZ71|A0A5C3PZ71_9APHY NAD(P)-binding protein OS=Polyporus arcularius HHB13444 OX=1314778 GN=K466DRAFT_596096 PE=3 SV=1
MM1 pKa = 7.82AEE3 pKa = 4.32LSVLTARR10 pKa = 11.84PSSSSRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84LPKK25 pKa = 10.19QVGAPLHH32 pKa = 6.12RR33 pKa = 11.84ASHH36 pKa = 5.65PRR38 pKa = 11.84PRR40 pKa = 11.84ALRR43 pKa = 11.84LPVRR47 pKa = 11.84HH48 pKa = 6.16LRR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84TPPRR57 pKa = 11.84PQLTPRR63 pKa = 11.84WPRR66 pKa = 11.84HH67 pKa = 5.05IPPPLLLPLRR77 pKa = 11.84HH78 pKa = 5.76RR79 pKa = 11.84QRR81 pKa = 11.84MQHH84 pKa = 4.37HH85 pKa = 5.67HH86 pKa = 6.09HH87 pKa = 6.09RR88 pKa = 11.84RR89 pKa = 11.84SRR91 pKa = 11.84RR92 pKa = 11.84VMWQTCSRR100 pKa = 11.84AALRR104 pKa = 11.84RR105 pKa = 11.84TGHH108 pKa = 4.55TTMCRR113 pKa = 11.84GYY115 pKa = 10.54LGSLVSTLRR124 pKa = 11.84HH125 pKa = 6.08LPPSLWLSPRR135 pKa = 11.84PHH137 pKa = 7.8HH138 pKa = 6.7GMQHH142 pKa = 4.82TRR144 pKa = 11.84HH145 pKa = 5.92PRR147 pKa = 11.84LRR149 pKa = 11.84LPRR152 pKa = 11.84LQRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84TGKK160 pKa = 8.99TSVCSSGTRR169 pKa = 11.84SGLSKK174 pKa = 10.47LLKK177 pKa = 9.65PRR179 pKa = 11.84RR180 pKa = 11.84PRR182 pKa = 3.63
MM1 pKa = 7.82AEE3 pKa = 4.32LSVLTARR10 pKa = 11.84PSSSSRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84LPKK25 pKa = 10.19QVGAPLHH32 pKa = 6.12RR33 pKa = 11.84ASHH36 pKa = 5.65PRR38 pKa = 11.84PRR40 pKa = 11.84ALRR43 pKa = 11.84LPVRR47 pKa = 11.84HH48 pKa = 6.16LRR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84TPPRR57 pKa = 11.84PQLTPRR63 pKa = 11.84WPRR66 pKa = 11.84HH67 pKa = 5.05IPPPLLLPLRR77 pKa = 11.84HH78 pKa = 5.76RR79 pKa = 11.84QRR81 pKa = 11.84MQHH84 pKa = 4.37HH85 pKa = 5.67HH86 pKa = 6.09HH87 pKa = 6.09RR88 pKa = 11.84RR89 pKa = 11.84SRR91 pKa = 11.84RR92 pKa = 11.84VMWQTCSRR100 pKa = 11.84AALRR104 pKa = 11.84RR105 pKa = 11.84TGHH108 pKa = 4.55TTMCRR113 pKa = 11.84GYY115 pKa = 10.54LGSLVSTLRR124 pKa = 11.84HH125 pKa = 6.08LPPSLWLSPRR135 pKa = 11.84PHH137 pKa = 7.8HH138 pKa = 6.7GMQHH142 pKa = 4.82TRR144 pKa = 11.84HH145 pKa = 5.92PRR147 pKa = 11.84LRR149 pKa = 11.84LPRR152 pKa = 11.84LQRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84TGKK160 pKa = 8.99TSVCSSGTRR169 pKa = 11.84SGLSKK174 pKa = 10.47LLKK177 pKa = 9.65PRR179 pKa = 11.84RR180 pKa = 11.84PRR182 pKa = 3.63
Molecular weight: 21.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6874114 |
49 |
5043 |
393.4 |
43.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.427 ± 0.018 | 1.397 ± 0.009 |
5.675 ± 0.012 | 5.784 ± 0.017 |
3.557 ± 0.014 | 6.534 ± 0.02 |
2.692 ± 0.009 | 4.291 ± 0.012 |
3.969 ± 0.017 | 9.244 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.057 ± 0.007 | 2.909 ± 0.01 |
6.9 ± 0.025 | 3.57 ± 0.01 |
6.907 ± 0.019 | 8.324 ± 0.024 |
5.967 ± 0.012 | 6.617 ± 0.012 |
1.519 ± 0.008 | 2.662 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
