
Phycisphaerae bacterium RAS2
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Phycisphaerae; unclassified Phycisphaerae
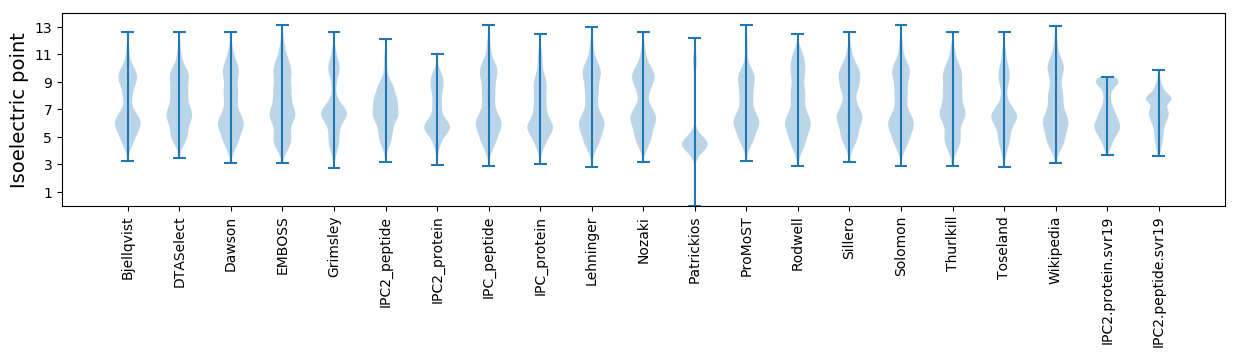
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
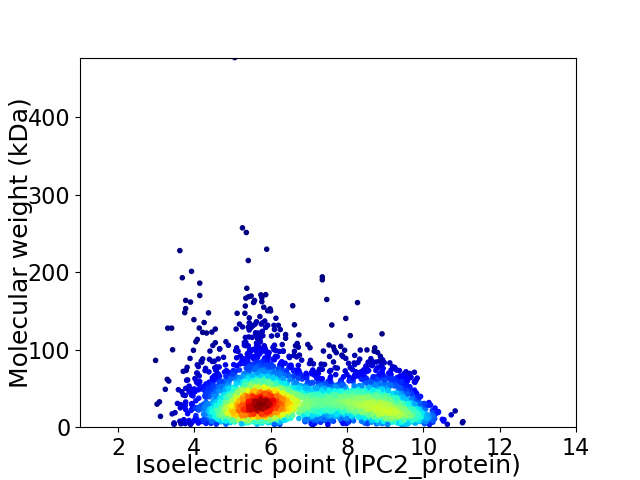
Virtual 2D-PAGE plot for 3554 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518LJG2|A0A518LJG2_9BACT UDP-2 3-diacetamido-2 3-dideoxy-D-glucuronate 2-epimerase OS=Phycisphaerae bacterium RAS2 OX=2528035 GN=wbpI_1 PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.76ALTWVIAGLLGVAVSASQAQTLYY26 pKa = 8.27TTGFDD31 pKa = 3.31GSPFVNGNLSGQDD44 pKa = 2.83GWVTTDD50 pKa = 3.11SPEE53 pKa = 4.13TPFLGTVQGSFSFSPSRR70 pKa = 11.84SVRR73 pKa = 11.84INASQTIDD81 pKa = 3.89SNWWWTPLNHH91 pKa = 6.9SIHH94 pKa = 6.54GSATVIQIEE103 pKa = 4.23CMIYY107 pKa = 9.73VDD109 pKa = 4.3RR110 pKa = 11.84ASGSASTLWGVDD122 pKa = 2.97IYY124 pKa = 11.42DD125 pKa = 3.74GSLPLTRR132 pKa = 11.84RR133 pKa = 11.84VATIAIDD140 pKa = 3.7TSNHH144 pKa = 5.22LLVWDD149 pKa = 4.07SVAFVDD155 pKa = 4.01TGVIVPPNTWGLFRR169 pKa = 11.84MNLNYY174 pKa = 10.48AAGEE178 pKa = 4.3RR179 pKa = 11.84NTSVYY184 pKa = 10.58FNGARR189 pKa = 11.84VAYY192 pKa = 9.82RR193 pKa = 11.84RR194 pKa = 11.84AFGLGANDD202 pKa = 3.31ILADD206 pKa = 3.36VDD208 pKa = 5.05FYY210 pKa = 11.6HH211 pKa = 7.54VDD213 pKa = 2.91AGGNDD218 pKa = 3.09SAYY221 pKa = 10.31FDD223 pKa = 3.97NFVVRR228 pKa = 11.84AMADD232 pKa = 3.2GDD234 pKa = 4.1GDD236 pKa = 4.38GVPDD240 pKa = 4.69SQDD243 pKa = 3.02SCLNTAAGDD252 pKa = 4.19PINATGCSTLDD263 pKa = 3.4NDD265 pKa = 5.34GDD267 pKa = 4.38GVLNDD272 pKa = 5.12ADD274 pKa = 4.26NCPDD278 pKa = 3.68TPTCAIVNGAGCPFDD293 pKa = 4.82SDD295 pKa = 3.73GDD297 pKa = 4.23SVMNGCDD304 pKa = 3.01NCDD307 pKa = 3.36NVPNGPGEE315 pKa = 4.63DD316 pKa = 3.67NQADD320 pKa = 3.83ADD322 pKa = 3.92SDD324 pKa = 4.04GRR326 pKa = 11.84GDD328 pKa = 3.76ACDD331 pKa = 3.23VCPFRR336 pKa = 11.84RR337 pKa = 11.84PGDD340 pKa = 3.56TTGDD344 pKa = 3.95GIVDD348 pKa = 3.52GRR350 pKa = 11.84DD351 pKa = 2.98AQLFVRR357 pKa = 11.84ILTGGSGTPDD367 pKa = 3.22EE368 pKa = 4.7TCACDD373 pKa = 3.57VNEE376 pKa = 4.06DD377 pKa = 4.08TFVNEE382 pKa = 3.83ADD384 pKa = 3.33IPALVTLLLNQQ395 pKa = 3.7
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.76ALTWVIAGLLGVAVSASQAQTLYY26 pKa = 8.27TTGFDD31 pKa = 3.31GSPFVNGNLSGQDD44 pKa = 2.83GWVTTDD50 pKa = 3.11SPEE53 pKa = 4.13TPFLGTVQGSFSFSPSRR70 pKa = 11.84SVRR73 pKa = 11.84INASQTIDD81 pKa = 3.89SNWWWTPLNHH91 pKa = 6.9SIHH94 pKa = 6.54GSATVIQIEE103 pKa = 4.23CMIYY107 pKa = 9.73VDD109 pKa = 4.3RR110 pKa = 11.84ASGSASTLWGVDD122 pKa = 2.97IYY124 pKa = 11.42DD125 pKa = 3.74GSLPLTRR132 pKa = 11.84RR133 pKa = 11.84VATIAIDD140 pKa = 3.7TSNHH144 pKa = 5.22LLVWDD149 pKa = 4.07SVAFVDD155 pKa = 4.01TGVIVPPNTWGLFRR169 pKa = 11.84MNLNYY174 pKa = 10.48AAGEE178 pKa = 4.3RR179 pKa = 11.84NTSVYY184 pKa = 10.58FNGARR189 pKa = 11.84VAYY192 pKa = 9.82RR193 pKa = 11.84RR194 pKa = 11.84AFGLGANDD202 pKa = 3.31ILADD206 pKa = 3.36VDD208 pKa = 5.05FYY210 pKa = 11.6HH211 pKa = 7.54VDD213 pKa = 2.91AGGNDD218 pKa = 3.09SAYY221 pKa = 10.31FDD223 pKa = 3.97NFVVRR228 pKa = 11.84AMADD232 pKa = 3.2GDD234 pKa = 4.1GDD236 pKa = 4.38GVPDD240 pKa = 4.69SQDD243 pKa = 3.02SCLNTAAGDD252 pKa = 4.19PINATGCSTLDD263 pKa = 3.4NDD265 pKa = 5.34GDD267 pKa = 4.38GVLNDD272 pKa = 5.12ADD274 pKa = 4.26NCPDD278 pKa = 3.68TPTCAIVNGAGCPFDD293 pKa = 4.82SDD295 pKa = 3.73GDD297 pKa = 4.23SVMNGCDD304 pKa = 3.01NCDD307 pKa = 3.36NVPNGPGEE315 pKa = 4.63DD316 pKa = 3.67NQADD320 pKa = 3.83ADD322 pKa = 3.92SDD324 pKa = 4.04GRR326 pKa = 11.84GDD328 pKa = 3.76ACDD331 pKa = 3.23VCPFRR336 pKa = 11.84RR337 pKa = 11.84PGDD340 pKa = 3.56TTGDD344 pKa = 3.95GIVDD348 pKa = 3.52GRR350 pKa = 11.84DD351 pKa = 2.98AQLFVRR357 pKa = 11.84ILTGGSGTPDD367 pKa = 3.22EE368 pKa = 4.7TCACDD373 pKa = 3.57VNEE376 pKa = 4.06DD377 pKa = 4.08TFVNEE382 pKa = 3.83ADD384 pKa = 3.33IPALVTLLLNQQ395 pKa = 3.7
Molecular weight: 41.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518LPJ3|A0A518LPJ3_9BACT ATPase family associated with various cellular activities (AAA) OS=Phycisphaerae bacterium RAS2 OX=2528035 GN=RAS2_29520 PE=4 SV=1
MM1 pKa = 7.78ISCQWPVISGQRR13 pKa = 11.84PLIGWHH19 pKa = 6.63GGGFWRR25 pKa = 11.84PSWSNVQPRR34 pKa = 11.84RR35 pKa = 11.84LVAGTDD41 pKa = 3.37SLIRR45 pKa = 11.84NATNGRR51 pKa = 11.84QSGVMRR57 pKa = 11.84FEE59 pKa = 4.67LLLPAARR66 pKa = 11.84MAIVIRR72 pKa = 11.84SVHH75 pKa = 4.87WLLRR79 pKa = 11.84RR80 pKa = 11.84LARR83 pKa = 11.84GRR85 pKa = 11.84INRR88 pKa = 11.84RR89 pKa = 11.84GLSAGARR96 pKa = 11.84SAAAVRR102 pKa = 11.84LGRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84GRR109 pKa = 11.84GIAIHH114 pKa = 6.59RR115 pKa = 11.84RR116 pKa = 11.84TGARR120 pKa = 11.84GHH122 pKa = 5.82EE123 pKa = 4.07VRR125 pKa = 11.84QRR127 pKa = 11.84GVEE130 pKa = 3.83IARR133 pKa = 11.84RR134 pKa = 11.84AHH136 pKa = 5.4AVWRR140 pKa = 11.84RR141 pKa = 11.84SVGSLRR147 pKa = 11.84TALARR152 pKa = 11.84RR153 pKa = 11.84SITRR157 pKa = 11.84RR158 pKa = 11.84SAGADD163 pKa = 2.7IASITGCRR171 pKa = 11.84RR172 pKa = 11.84VARR175 pKa = 11.84RR176 pKa = 11.84GSIPRR181 pKa = 11.84WRR183 pKa = 11.84TISIAA188 pKa = 3.3
MM1 pKa = 7.78ISCQWPVISGQRR13 pKa = 11.84PLIGWHH19 pKa = 6.63GGGFWRR25 pKa = 11.84PSWSNVQPRR34 pKa = 11.84RR35 pKa = 11.84LVAGTDD41 pKa = 3.37SLIRR45 pKa = 11.84NATNGRR51 pKa = 11.84QSGVMRR57 pKa = 11.84FEE59 pKa = 4.67LLLPAARR66 pKa = 11.84MAIVIRR72 pKa = 11.84SVHH75 pKa = 4.87WLLRR79 pKa = 11.84RR80 pKa = 11.84LARR83 pKa = 11.84GRR85 pKa = 11.84INRR88 pKa = 11.84RR89 pKa = 11.84GLSAGARR96 pKa = 11.84SAAAVRR102 pKa = 11.84LGRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84GRR109 pKa = 11.84GIAIHH114 pKa = 6.59RR115 pKa = 11.84RR116 pKa = 11.84TGARR120 pKa = 11.84GHH122 pKa = 5.82EE123 pKa = 4.07VRR125 pKa = 11.84QRR127 pKa = 11.84GVEE130 pKa = 3.83IARR133 pKa = 11.84RR134 pKa = 11.84AHH136 pKa = 5.4AVWRR140 pKa = 11.84RR141 pKa = 11.84SVGSLRR147 pKa = 11.84TALARR152 pKa = 11.84RR153 pKa = 11.84SITRR157 pKa = 11.84RR158 pKa = 11.84SAGADD163 pKa = 2.7IASITGCRR171 pKa = 11.84RR172 pKa = 11.84VARR175 pKa = 11.84RR176 pKa = 11.84GSIPRR181 pKa = 11.84WRR183 pKa = 11.84TISIAA188 pKa = 3.3
Molecular weight: 21.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1265709 |
29 |
4444 |
356.1 |
38.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.575 ± 0.053 | 1.337 ± 0.023 |
5.839 ± 0.035 | 5.579 ± 0.042 |
3.594 ± 0.026 | 8.212 ± 0.046 |
2.135 ± 0.019 | 4.934 ± 0.026 |
3.342 ± 0.039 | 9.57 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.348 ± 0.017 | 2.944 ± 0.033 |
5.463 ± 0.031 | 3.293 ± 0.022 |
7.579 ± 0.051 | 5.744 ± 0.027 |
5.43 ± 0.036 | 7.383 ± 0.033 |
1.409 ± 0.02 | 2.29 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
