
Sphingomonas sp. CL5.1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
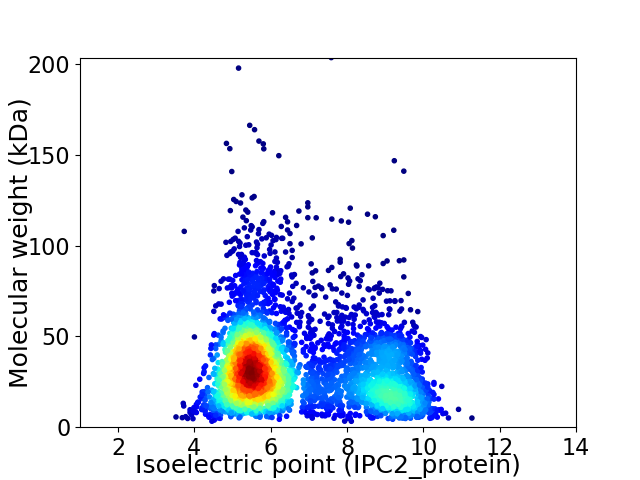
Virtual 2D-PAGE plot for 4226 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N0WWD1|A0A6N0WWD1_9SPHN Cytochrome P450 OS=Sphingomonas sp. CL5.1 OX=2653203 GN=F9288_00855 PE=3 SV=1
MM1 pKa = 7.62SNTTSVGINLSPVNYY16 pKa = 9.43WSTEE20 pKa = 3.81YY21 pKa = 10.61PFLDD25 pKa = 3.21RR26 pKa = 11.84MKK28 pKa = 10.43TAGTWGVQGTSVSSLIVDD46 pKa = 3.38QNGYY50 pKa = 6.9PTTIPTGATSVSTIIALDD68 pKa = 3.77PASAGTDD75 pKa = 3.1NTYY78 pKa = 11.19VLTYY82 pKa = 9.98TGTATFRR89 pKa = 11.84IPGATIVSSKK99 pKa = 10.06PGEE102 pKa = 3.78ITFTVTGANPMQTVMITSIAASNPLTAMHH131 pKa = 6.67VVRR134 pKa = 11.84QDD136 pKa = 3.08QMALFNQGEE145 pKa = 4.54LFNPAFVKK153 pKa = 10.42QVSQFGTLRR162 pKa = 11.84YY163 pKa = 9.06MDD165 pKa = 3.95WEE167 pKa = 4.19QTNITTATSWSQRR180 pKa = 11.84TTPDD184 pKa = 3.15TLSWASTGSTTGVPIEE200 pKa = 4.6AMVALANEE208 pKa = 4.48SRR210 pKa = 11.84TNMWLNIPTQANDD223 pKa = 3.44DD224 pKa = 4.19YY225 pKa = 10.74VRR227 pKa = 11.84QMLTYY232 pKa = 10.6VRR234 pKa = 11.84DD235 pKa = 3.85NLDD238 pKa = 3.28PSLKK242 pKa = 10.86VNVEE246 pKa = 4.06YY247 pKa = 11.17SNEE250 pKa = 3.45MWNWGFQQSRR260 pKa = 11.84YY261 pKa = 10.09ANQLATQLWGTNVTRR276 pKa = 11.84GVQQYY281 pKa = 9.6YY282 pKa = 10.54GYY284 pKa = 10.77RR285 pKa = 11.84SAQIAAIANDD295 pKa = 3.56VFGDD299 pKa = 4.58GAATRR304 pKa = 11.84LDD306 pKa = 3.73NVLATQTANLGLEE319 pKa = 4.04KK320 pKa = 11.0YY321 pKa = 10.08IFEE324 pKa = 4.59GVAKK328 pKa = 10.5AGLGDD333 pKa = 3.71VSSLFQSYY341 pKa = 10.53AVTTYY346 pKa = 10.45FGSEE350 pKa = 3.64LSGQNSADD358 pKa = 3.87RR359 pKa = 11.84ATILGWAKK367 pKa = 10.49GGSAGLDD374 pKa = 3.0AAFNEE379 pKa = 4.5IANGGTLTGWGSLPSIIAQWAYY401 pKa = 9.78QATVAKK407 pKa = 10.27KK408 pKa = 10.18YY409 pKa = 10.87GLDD412 pKa = 3.26LVAYY416 pKa = 9.0EE417 pKa = 4.54GGIDD421 pKa = 3.77LSAAGFSAADD431 pKa = 3.42QPIVMDD437 pKa = 3.85FFSRR441 pKa = 11.84LEE443 pKa = 4.04ADD445 pKa = 3.22PRR447 pKa = 11.84MATIYY452 pKa = 9.78TQMIDD457 pKa = 3.19SFSAAGGTLLNAYY470 pKa = 8.71TDD472 pKa = 3.88AQVDD476 pKa = 4.2RR477 pKa = 11.84PGGLYY482 pKa = 8.52GTLKK486 pKa = 10.51SIYY489 pKa = 10.19DD490 pKa = 3.72SASPAWSAPVNGQAEE505 pKa = 4.3ASKK508 pKa = 11.14ASGPVPANNVSGLPAANQPTPVAQNITTAASSYY541 pKa = 8.04TLEE544 pKa = 4.53SGAMTLSYY552 pKa = 10.01IGQSGFVGTGNDD564 pKa = 3.79LDD566 pKa = 4.11NVITGGNSGNKK577 pKa = 10.0LYY579 pKa = 11.06GGAGNDD585 pKa = 3.77TLIGGAGNDD594 pKa = 3.79YY595 pKa = 11.17LDD597 pKa = 4.62GGTGADD603 pKa = 3.73TMTGGAGNDD612 pKa = 3.12IYY614 pKa = 10.92IVDD617 pKa = 3.85NPGDD621 pKa = 3.97VVVEE625 pKa = 4.13AANGGTDD632 pKa = 3.54EE633 pKa = 4.57VRR635 pKa = 11.84TSLDD639 pKa = 3.4SYY641 pKa = 10.79TLGANIEE648 pKa = 4.16NLTYY652 pKa = 9.9TGSGAFTGTGNDD664 pKa = 3.55LANVITGGDD673 pKa = 3.99GGNKK677 pKa = 10.1LYY679 pKa = 10.73GYY681 pKa = 10.05DD682 pKa = 4.74GNDD685 pKa = 3.4TLIGGAGNDD694 pKa = 3.79YY695 pKa = 11.17LDD697 pKa = 4.56GGTGADD703 pKa = 3.44RR704 pKa = 11.84MVGGAGNDD712 pKa = 3.53TYY714 pKa = 11.17IVDD717 pKa = 3.73NPGDD721 pKa = 3.85VVVEE725 pKa = 4.01LPGGGIDD732 pKa = 4.25EE733 pKa = 4.59VRR735 pKa = 11.84TSLGSYY741 pKa = 10.57ALGANLEE748 pKa = 4.21NLTATGSSTFTGIGNAADD766 pKa = 3.78NVITGSTLGANKK778 pKa = 10.49LYY780 pKa = 10.69GYY782 pKa = 10.12DD783 pKa = 4.45GNDD786 pKa = 3.4TLIGGAGNDD795 pKa = 3.79YY796 pKa = 11.17LDD798 pKa = 4.36GGTGADD804 pKa = 3.82YY805 pKa = 9.51MAGGDD810 pKa = 3.73GDD812 pKa = 3.84DD813 pKa = 4.28TYY815 pKa = 12.05VVDD818 pKa = 3.83NPGDD822 pKa = 3.77VVVEE826 pKa = 4.01LPGGGTDD833 pKa = 3.51TVVSSISYY841 pKa = 8.76TLGANLEE848 pKa = 3.95NLRR851 pKa = 11.84LGGSGNINGAGNDD864 pKa = 3.44AANIITGNAGNNILSGGGGDD884 pKa = 5.44DD885 pKa = 3.61VLQGGDD891 pKa = 3.55GDD893 pKa = 5.12DD894 pKa = 4.04YY895 pKa = 11.89LDD897 pKa = 4.62GGDD900 pKa = 4.74GNDD903 pKa = 3.96SLSGGNGNDD912 pKa = 3.2ILIGGAGNDD921 pKa = 3.76YY922 pKa = 11.12LSGGNGNDD930 pKa = 3.5VLIGGPGKK938 pKa = 10.4DD939 pKa = 3.55SLVGGAGADD948 pKa = 2.92RR949 pKa = 11.84FVFAPGDD956 pKa = 3.67LGNTITNTDD965 pKa = 3.76TILDD969 pKa = 4.18FSHH972 pKa = 7.49AEE974 pKa = 3.66GDD976 pKa = 4.82KK977 pKa = 10.6IDD979 pKa = 5.17LSQFDD984 pKa = 4.93ANTNTIAKK992 pKa = 9.96DD993 pKa = 3.16AFTFIANKK1001 pKa = 9.55PFSHH1005 pKa = 6.56TAGEE1009 pKa = 4.22VRR1011 pKa = 11.84VDD1013 pKa = 4.26SIGGYY1018 pKa = 7.77WNVMGDD1024 pKa = 3.64TNGDD1028 pKa = 3.37GVADD1032 pKa = 4.49FALNVKK1038 pKa = 10.46ASGPLVQSDD1047 pKa = 4.87FILGG1051 pKa = 3.53
MM1 pKa = 7.62SNTTSVGINLSPVNYY16 pKa = 9.43WSTEE20 pKa = 3.81YY21 pKa = 10.61PFLDD25 pKa = 3.21RR26 pKa = 11.84MKK28 pKa = 10.43TAGTWGVQGTSVSSLIVDD46 pKa = 3.38QNGYY50 pKa = 6.9PTTIPTGATSVSTIIALDD68 pKa = 3.77PASAGTDD75 pKa = 3.1NTYY78 pKa = 11.19VLTYY82 pKa = 9.98TGTATFRR89 pKa = 11.84IPGATIVSSKK99 pKa = 10.06PGEE102 pKa = 3.78ITFTVTGANPMQTVMITSIAASNPLTAMHH131 pKa = 6.67VVRR134 pKa = 11.84QDD136 pKa = 3.08QMALFNQGEE145 pKa = 4.54LFNPAFVKK153 pKa = 10.42QVSQFGTLRR162 pKa = 11.84YY163 pKa = 9.06MDD165 pKa = 3.95WEE167 pKa = 4.19QTNITTATSWSQRR180 pKa = 11.84TTPDD184 pKa = 3.15TLSWASTGSTTGVPIEE200 pKa = 4.6AMVALANEE208 pKa = 4.48SRR210 pKa = 11.84TNMWLNIPTQANDD223 pKa = 3.44DD224 pKa = 4.19YY225 pKa = 10.74VRR227 pKa = 11.84QMLTYY232 pKa = 10.6VRR234 pKa = 11.84DD235 pKa = 3.85NLDD238 pKa = 3.28PSLKK242 pKa = 10.86VNVEE246 pKa = 4.06YY247 pKa = 11.17SNEE250 pKa = 3.45MWNWGFQQSRR260 pKa = 11.84YY261 pKa = 10.09ANQLATQLWGTNVTRR276 pKa = 11.84GVQQYY281 pKa = 9.6YY282 pKa = 10.54GYY284 pKa = 10.77RR285 pKa = 11.84SAQIAAIANDD295 pKa = 3.56VFGDD299 pKa = 4.58GAATRR304 pKa = 11.84LDD306 pKa = 3.73NVLATQTANLGLEE319 pKa = 4.04KK320 pKa = 11.0YY321 pKa = 10.08IFEE324 pKa = 4.59GVAKK328 pKa = 10.5AGLGDD333 pKa = 3.71VSSLFQSYY341 pKa = 10.53AVTTYY346 pKa = 10.45FGSEE350 pKa = 3.64LSGQNSADD358 pKa = 3.87RR359 pKa = 11.84ATILGWAKK367 pKa = 10.49GGSAGLDD374 pKa = 3.0AAFNEE379 pKa = 4.5IANGGTLTGWGSLPSIIAQWAYY401 pKa = 9.78QATVAKK407 pKa = 10.27KK408 pKa = 10.18YY409 pKa = 10.87GLDD412 pKa = 3.26LVAYY416 pKa = 9.0EE417 pKa = 4.54GGIDD421 pKa = 3.77LSAAGFSAADD431 pKa = 3.42QPIVMDD437 pKa = 3.85FFSRR441 pKa = 11.84LEE443 pKa = 4.04ADD445 pKa = 3.22PRR447 pKa = 11.84MATIYY452 pKa = 9.78TQMIDD457 pKa = 3.19SFSAAGGTLLNAYY470 pKa = 8.71TDD472 pKa = 3.88AQVDD476 pKa = 4.2RR477 pKa = 11.84PGGLYY482 pKa = 8.52GTLKK486 pKa = 10.51SIYY489 pKa = 10.19DD490 pKa = 3.72SASPAWSAPVNGQAEE505 pKa = 4.3ASKK508 pKa = 11.14ASGPVPANNVSGLPAANQPTPVAQNITTAASSYY541 pKa = 8.04TLEE544 pKa = 4.53SGAMTLSYY552 pKa = 10.01IGQSGFVGTGNDD564 pKa = 3.79LDD566 pKa = 4.11NVITGGNSGNKK577 pKa = 10.0LYY579 pKa = 11.06GGAGNDD585 pKa = 3.77TLIGGAGNDD594 pKa = 3.79YY595 pKa = 11.17LDD597 pKa = 4.62GGTGADD603 pKa = 3.73TMTGGAGNDD612 pKa = 3.12IYY614 pKa = 10.92IVDD617 pKa = 3.85NPGDD621 pKa = 3.97VVVEE625 pKa = 4.13AANGGTDD632 pKa = 3.54EE633 pKa = 4.57VRR635 pKa = 11.84TSLDD639 pKa = 3.4SYY641 pKa = 10.79TLGANIEE648 pKa = 4.16NLTYY652 pKa = 9.9TGSGAFTGTGNDD664 pKa = 3.55LANVITGGDD673 pKa = 3.99GGNKK677 pKa = 10.1LYY679 pKa = 10.73GYY681 pKa = 10.05DD682 pKa = 4.74GNDD685 pKa = 3.4TLIGGAGNDD694 pKa = 3.79YY695 pKa = 11.17LDD697 pKa = 4.56GGTGADD703 pKa = 3.44RR704 pKa = 11.84MVGGAGNDD712 pKa = 3.53TYY714 pKa = 11.17IVDD717 pKa = 3.73NPGDD721 pKa = 3.85VVVEE725 pKa = 4.01LPGGGIDD732 pKa = 4.25EE733 pKa = 4.59VRR735 pKa = 11.84TSLGSYY741 pKa = 10.57ALGANLEE748 pKa = 4.21NLTATGSSTFTGIGNAADD766 pKa = 3.78NVITGSTLGANKK778 pKa = 10.49LYY780 pKa = 10.69GYY782 pKa = 10.12DD783 pKa = 4.45GNDD786 pKa = 3.4TLIGGAGNDD795 pKa = 3.79YY796 pKa = 11.17LDD798 pKa = 4.36GGTGADD804 pKa = 3.82YY805 pKa = 9.51MAGGDD810 pKa = 3.73GDD812 pKa = 3.84DD813 pKa = 4.28TYY815 pKa = 12.05VVDD818 pKa = 3.83NPGDD822 pKa = 3.77VVVEE826 pKa = 4.01LPGGGTDD833 pKa = 3.51TVVSSISYY841 pKa = 8.76TLGANLEE848 pKa = 3.95NLRR851 pKa = 11.84LGGSGNINGAGNDD864 pKa = 3.44AANIITGNAGNNILSGGGGDD884 pKa = 5.44DD885 pKa = 3.61VLQGGDD891 pKa = 3.55GDD893 pKa = 5.12DD894 pKa = 4.04YY895 pKa = 11.89LDD897 pKa = 4.62GGDD900 pKa = 4.74GNDD903 pKa = 3.96SLSGGNGNDD912 pKa = 3.2ILIGGAGNDD921 pKa = 3.76YY922 pKa = 11.12LSGGNGNDD930 pKa = 3.5VLIGGPGKK938 pKa = 10.4DD939 pKa = 3.55SLVGGAGADD948 pKa = 2.92RR949 pKa = 11.84FVFAPGDD956 pKa = 3.67LGNTITNTDD965 pKa = 3.76TILDD969 pKa = 4.18FSHH972 pKa = 7.49AEE974 pKa = 3.66GDD976 pKa = 4.82KK977 pKa = 10.6IDD979 pKa = 5.17LSQFDD984 pKa = 4.93ANTNTIAKK992 pKa = 9.96DD993 pKa = 3.16AFTFIANKK1001 pKa = 9.55PFSHH1005 pKa = 6.56TAGEE1009 pKa = 4.22VRR1011 pKa = 11.84VDD1013 pKa = 4.26SIGGYY1018 pKa = 7.77WNVMGDD1024 pKa = 3.64TNGDD1028 pKa = 3.37GVADD1032 pKa = 4.49FALNVKK1038 pKa = 10.46ASGPLVQSDD1047 pKa = 4.87FILGG1051 pKa = 3.53
Molecular weight: 107.89 kDa
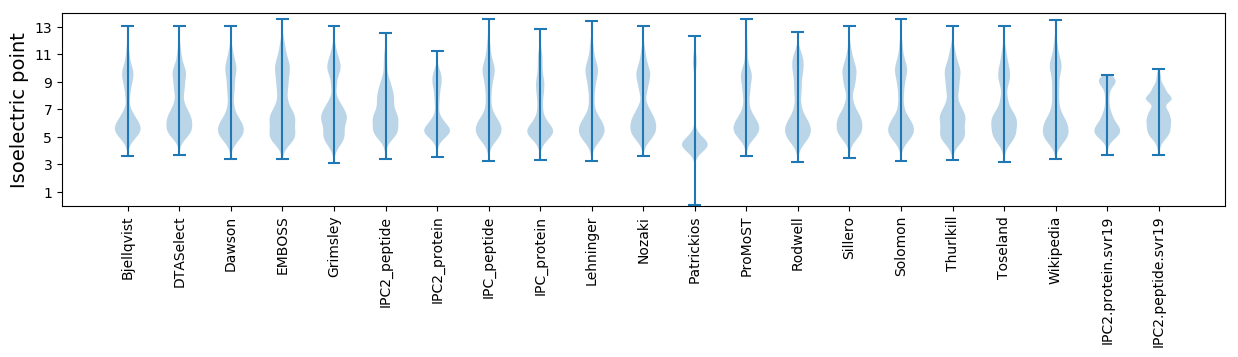
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N0X6D9|A0A6N0X6D9_9SPHN DUF1778 domain-containing protein OS=Sphingomonas sp. CL5.1 OX=2653203 GN=F9288_01675 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1378064 |
29 |
1920 |
326.1 |
35.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.036 ± 0.065 | 0.759 ± 0.011 |
5.929 ± 0.029 | 5.198 ± 0.037 |
3.495 ± 0.021 | 9.103 ± 0.04 |
2.015 ± 0.017 | 4.989 ± 0.022 |
2.654 ± 0.027 | 9.719 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.018 | 2.461 ± 0.028 |
5.446 ± 0.025 | 2.827 ± 0.022 |
7.791 ± 0.036 | 4.913 ± 0.033 |
5.283 ± 0.029 | 7.303 ± 0.032 |
1.444 ± 0.015 | 2.28 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
