
Pyrrhoderma noxium
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Hymenochaetales; Hymenochaetaceae; Pyrrhoderma
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
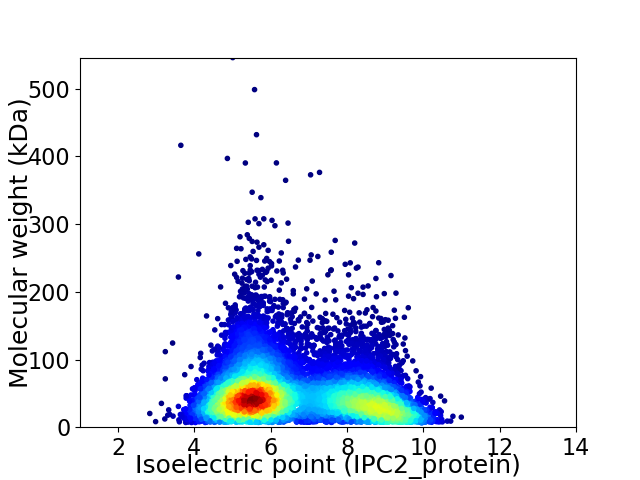
Virtual 2D-PAGE plot for 9911 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
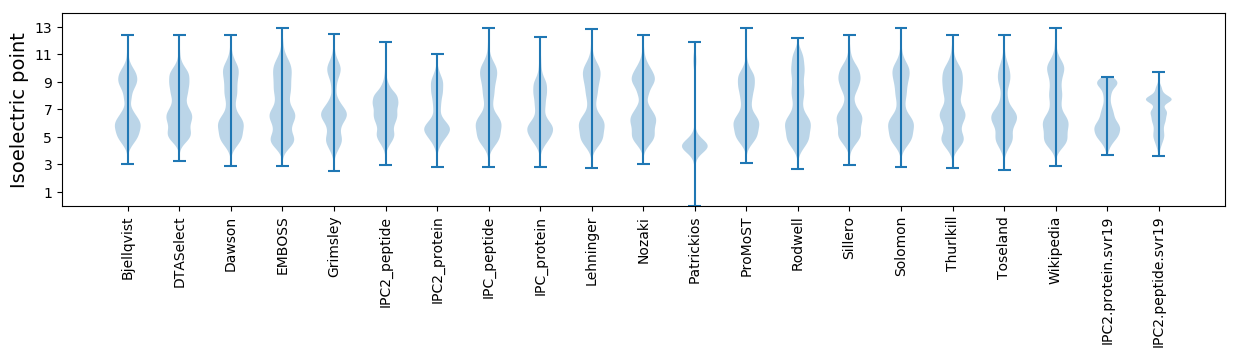
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286UT56|A0A286UT56_9AGAM Glycosyltransferase family 15 OS=Pyrrhoderma noxium OX=2282107 GN=PNOK_0259200 PE=3 SV=1
MM1 pKa = 7.94IIRR4 pKa = 11.84NGIRR8 pKa = 11.84DD9 pKa = 3.74AFLYY13 pKa = 11.05LLGALFILSRR23 pKa = 11.84VEE25 pKa = 4.15AYY27 pKa = 10.72VPALPSNDD35 pKa = 2.96STLIEE40 pKa = 4.19TSVNQSDD47 pKa = 3.84TSRR50 pKa = 11.84LVLQWFPGSQFSQFVSYY67 pKa = 10.61QLAGADD73 pKa = 3.51STGVNMGALVHH84 pKa = 7.13FSEE87 pKa = 5.26QDD89 pKa = 3.09KK90 pKa = 10.72TNQTTDD96 pKa = 3.68TPWIALVACDD106 pKa = 4.34TNGTAYY112 pKa = 10.9SLEE115 pKa = 4.47DD116 pKa = 4.38DD117 pKa = 4.35VFTLARR123 pKa = 11.84DD124 pKa = 3.74RR125 pKa = 11.84GAVAALLYY133 pKa = 10.59SEE135 pKa = 5.2WSDD138 pKa = 3.13ACIINAEE145 pKa = 4.21YY146 pKa = 10.88ADD148 pKa = 4.69PSTFDD153 pKa = 3.02QVFDD157 pKa = 3.43IFSTKK162 pKa = 9.61TLLSAQTTEE171 pKa = 4.04NAFTNVNQTLYY182 pKa = 10.81SQFDD186 pKa = 3.55ATRR189 pKa = 11.84LNASLTNISNSITTGNVFPGYY210 pKa = 10.62LFATLTTANATVSQSGTDD228 pKa = 3.5SDD230 pKa = 4.48PSSSGSGSSSNGNTDD245 pKa = 2.99LAMIVLYY252 pKa = 10.43AITGAVSVLFCIVILSPTEE271 pKa = 3.54YY272 pKa = 10.67DD273 pKa = 3.25IIVPTSPRR281 pKa = 11.84SKK283 pKa = 10.3VSQDD287 pKa = 3.0PTTSLIQILVSNTSS301 pKa = 2.89
MM1 pKa = 7.94IIRR4 pKa = 11.84NGIRR8 pKa = 11.84DD9 pKa = 3.74AFLYY13 pKa = 11.05LLGALFILSRR23 pKa = 11.84VEE25 pKa = 4.15AYY27 pKa = 10.72VPALPSNDD35 pKa = 2.96STLIEE40 pKa = 4.19TSVNQSDD47 pKa = 3.84TSRR50 pKa = 11.84LVLQWFPGSQFSQFVSYY67 pKa = 10.61QLAGADD73 pKa = 3.51STGVNMGALVHH84 pKa = 7.13FSEE87 pKa = 5.26QDD89 pKa = 3.09KK90 pKa = 10.72TNQTTDD96 pKa = 3.68TPWIALVACDD106 pKa = 4.34TNGTAYY112 pKa = 10.9SLEE115 pKa = 4.47DD116 pKa = 4.38DD117 pKa = 4.35VFTLARR123 pKa = 11.84DD124 pKa = 3.74RR125 pKa = 11.84GAVAALLYY133 pKa = 10.59SEE135 pKa = 5.2WSDD138 pKa = 3.13ACIINAEE145 pKa = 4.21YY146 pKa = 10.88ADD148 pKa = 4.69PSTFDD153 pKa = 3.02QVFDD157 pKa = 3.43IFSTKK162 pKa = 9.61TLLSAQTTEE171 pKa = 4.04NAFTNVNQTLYY182 pKa = 10.81SQFDD186 pKa = 3.55ATRR189 pKa = 11.84LNASLTNISNSITTGNVFPGYY210 pKa = 10.62LFATLTTANATVSQSGTDD228 pKa = 3.5SDD230 pKa = 4.48PSSSGSGSSSNGNTDD245 pKa = 2.99LAMIVLYY252 pKa = 10.43AITGAVSVLFCIVILSPTEE271 pKa = 3.54YY272 pKa = 10.67DD273 pKa = 3.25IIVPTSPRR281 pKa = 11.84SKK283 pKa = 10.3VSQDD287 pKa = 3.0PTTSLIQILVSNTSS301 pKa = 2.89
Molecular weight: 32.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286UWP8|A0A286UWP8_9AGAM Uncharacterized protein OS=Pyrrhoderma noxium OX=2282107 GN=PNOK_0110100 PE=4 SV=1
MM1 pKa = 7.15GATWEE6 pKa = 4.23HH7 pKa = 6.39LKK9 pKa = 10.95DD10 pKa = 3.62DD11 pKa = 4.79AEE13 pKa = 4.22WKK15 pKa = 10.31EE16 pKa = 4.02LDD18 pKa = 3.39RR19 pKa = 11.84QRR21 pKa = 11.84LEE23 pKa = 4.68RR24 pKa = 11.84GEE26 pKa = 4.14QPASRR31 pKa = 11.84ISSIRR36 pKa = 11.84SSSSPDD42 pKa = 2.9QRR44 pKa = 11.84QPSTPANQRR53 pKa = 11.84PSHH56 pKa = 6.68ASPSIAQRR64 pKa = 11.84PPPISAPQRR73 pKa = 11.84QVPPPATQRR82 pKa = 11.84QPAYY86 pKa = 8.33PAHH89 pKa = 7.57PIQQPRR95 pKa = 11.84STTSASVGIATNSSARR111 pKa = 11.84LASTATYY118 pKa = 10.69SSGNRR123 pKa = 11.84APEE126 pKa = 3.94AAEE129 pKa = 3.99ATSSSSASDD138 pKa = 3.41KK139 pKa = 10.75TCEE142 pKa = 3.99LCHH145 pKa = 6.68KK146 pKa = 10.12RR147 pKa = 11.84FTRR150 pKa = 11.84TSDD153 pKa = 3.2LTRR156 pKa = 11.84HH157 pKa = 5.69IDD159 pKa = 3.15HH160 pKa = 7.31RR161 pKa = 11.84KK162 pKa = 6.73TLTVVTQCQRR172 pKa = 11.84YY173 pKa = 8.62RR174 pKa = 11.84RR175 pKa = 3.87
MM1 pKa = 7.15GATWEE6 pKa = 4.23HH7 pKa = 6.39LKK9 pKa = 10.95DD10 pKa = 3.62DD11 pKa = 4.79AEE13 pKa = 4.22WKK15 pKa = 10.31EE16 pKa = 4.02LDD18 pKa = 3.39RR19 pKa = 11.84QRR21 pKa = 11.84LEE23 pKa = 4.68RR24 pKa = 11.84GEE26 pKa = 4.14QPASRR31 pKa = 11.84ISSIRR36 pKa = 11.84SSSSPDD42 pKa = 2.9QRR44 pKa = 11.84QPSTPANQRR53 pKa = 11.84PSHH56 pKa = 6.68ASPSIAQRR64 pKa = 11.84PPPISAPQRR73 pKa = 11.84QVPPPATQRR82 pKa = 11.84QPAYY86 pKa = 8.33PAHH89 pKa = 7.57PIQQPRR95 pKa = 11.84STTSASVGIATNSSARR111 pKa = 11.84LASTATYY118 pKa = 10.69SSGNRR123 pKa = 11.84APEE126 pKa = 3.94AAEE129 pKa = 3.99ATSSSSASDD138 pKa = 3.41KK139 pKa = 10.75TCEE142 pKa = 3.99LCHH145 pKa = 6.68KK146 pKa = 10.12RR147 pKa = 11.84FTRR150 pKa = 11.84TSDD153 pKa = 3.2LTRR156 pKa = 11.84HH157 pKa = 5.69IDD159 pKa = 3.15HH160 pKa = 7.31RR161 pKa = 11.84KK162 pKa = 6.73TLTVVTQCQRR172 pKa = 11.84YY173 pKa = 8.62RR174 pKa = 11.84RR175 pKa = 3.87
Molecular weight: 19.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4934286 |
52 |
4846 |
497.9 |
55.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.165 ± 0.022 | 1.155 ± 0.009 |
5.535 ± 0.016 | 6.271 ± 0.027 |
3.758 ± 0.015 | 6.514 ± 0.031 |
2.311 ± 0.011 | 5.449 ± 0.023 |
5.086 ± 0.023 | 9.007 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.921 ± 0.009 | 4.201 ± 0.012 |
5.926 ± 0.03 | 3.642 ± 0.021 |
5.997 ± 0.021 | 9.803 ± 0.032 |
6.167 ± 0.02 | 6.006 ± 0.017 |
1.281 ± 0.009 | 2.805 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
