
Phellinidium pouzarii
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Hymenochaetales; Hymenochaetaceae; Phellinidium
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
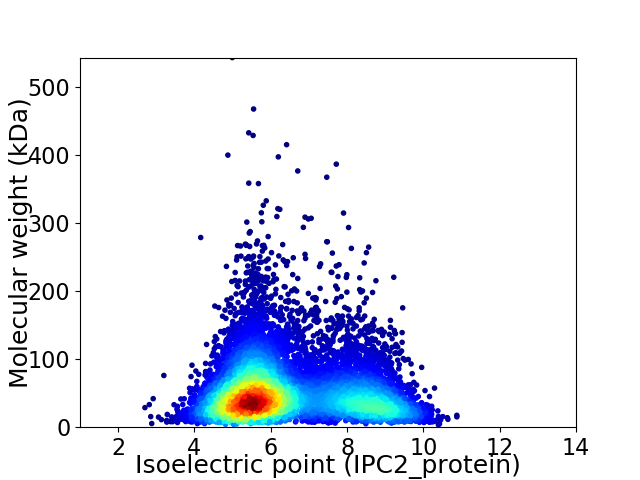
Virtual 2D-PAGE plot for 8697 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S4L1Y3|A0A4S4L1Y3_9AGAM Uncharacterized protein OS=Phellinidium pouzarii OX=167371 GN=EW145_g4877 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.48LSITFCVSTLVASTAAAPLDD22 pKa = 3.94LFWRR26 pKa = 11.84QVVADD31 pKa = 4.88GLTLTQIDD39 pKa = 4.27ALTPQFGHH47 pKa = 6.83AADD50 pKa = 4.04VNPTGTGNCDD60 pKa = 2.86GAVNDD65 pKa = 4.45TNGLPIEE72 pKa = 4.8VPCSCPPPRR81 pKa = 11.84ATFIQSLAANIAAGHH96 pKa = 6.03AVNNPSVAVTFPTDD110 pKa = 2.93NSTASILARR119 pKa = 11.84FNAASITLQNEE130 pKa = 3.59NGEE133 pKa = 4.39GVGCPEE139 pKa = 5.72SSTTWTAQAAAIQAGAKK156 pKa = 9.98DD157 pKa = 4.06NNAPTAAAPPSSSAPVSASSPATTVAAPASSAVTTASASAPSTTGADD204 pKa = 3.79GLSLAQIDD212 pKa = 4.2ALTPQFGHH220 pKa = 7.14DD221 pKa = 3.51ADD223 pKa = 4.28VNPTGTGNCDD233 pKa = 3.29GAVDD237 pKa = 4.58DD238 pKa = 5.72ANGDD242 pKa = 4.37PILVPCDD249 pKa = 3.58CPPPRR254 pKa = 11.84NQFIQDD260 pKa = 3.83LTANIAAGFAVNNPSVAITFPTDD283 pKa = 2.67NSTNSILARR292 pKa = 11.84FNAASVTLQNTNGPGVGCPISSTTWTAQANAIRR325 pKa = 11.84DD326 pKa = 4.02GQQSRR331 pKa = 11.84VAIATTLSVSTEE343 pKa = 4.21SSVSTSPAASSVVAGCPAPEE363 pKa = 4.07PTPTSSEE370 pKa = 4.16VPTPSSIVSASPVSSTDD387 pKa = 3.01VTSADD392 pKa = 3.92GLSLAQIDD400 pKa = 4.3ALTPRR405 pKa = 11.84LGFSSGVNPTGTGNCDD421 pKa = 3.09GAVNGANGQPIEE433 pKa = 4.7VPCSCPPARR442 pKa = 11.84DD443 pKa = 3.56VFIQSLAQDD452 pKa = 3.83IAAGHH457 pKa = 6.21AVNNPSVSVSFPTDD471 pKa = 2.78NSTSSQITRR480 pKa = 11.84IQSSLVALQNLNGPGQGCPAASTTLSAQLAALQGG514 pKa = 3.57
MM1 pKa = 7.46KK2 pKa = 10.48LSITFCVSTLVASTAAAPLDD22 pKa = 3.94LFWRR26 pKa = 11.84QVVADD31 pKa = 4.88GLTLTQIDD39 pKa = 4.27ALTPQFGHH47 pKa = 6.83AADD50 pKa = 4.04VNPTGTGNCDD60 pKa = 2.86GAVNDD65 pKa = 4.45TNGLPIEE72 pKa = 4.8VPCSCPPPRR81 pKa = 11.84ATFIQSLAANIAAGHH96 pKa = 6.03AVNNPSVAVTFPTDD110 pKa = 2.93NSTASILARR119 pKa = 11.84FNAASITLQNEE130 pKa = 3.59NGEE133 pKa = 4.39GVGCPEE139 pKa = 5.72SSTTWTAQAAAIQAGAKK156 pKa = 9.98DD157 pKa = 4.06NNAPTAAAPPSSSAPVSASSPATTVAAPASSAVTTASASAPSTTGADD204 pKa = 3.79GLSLAQIDD212 pKa = 4.2ALTPQFGHH220 pKa = 7.14DD221 pKa = 3.51ADD223 pKa = 4.28VNPTGTGNCDD233 pKa = 3.29GAVDD237 pKa = 4.58DD238 pKa = 5.72ANGDD242 pKa = 4.37PILVPCDD249 pKa = 3.58CPPPRR254 pKa = 11.84NQFIQDD260 pKa = 3.83LTANIAAGFAVNNPSVAITFPTDD283 pKa = 2.67NSTNSILARR292 pKa = 11.84FNAASVTLQNTNGPGVGCPISSTTWTAQANAIRR325 pKa = 11.84DD326 pKa = 4.02GQQSRR331 pKa = 11.84VAIATTLSVSTEE343 pKa = 4.21SSVSTSPAASSVVAGCPAPEE363 pKa = 4.07PTPTSSEE370 pKa = 4.16VPTPSSIVSASPVSSTDD387 pKa = 3.01VTSADD392 pKa = 3.92GLSLAQIDD400 pKa = 4.3ALTPRR405 pKa = 11.84LGFSSGVNPTGTGNCDD421 pKa = 3.09GAVNGANGQPIEE433 pKa = 4.7VPCSCPPARR442 pKa = 11.84DD443 pKa = 3.56VFIQSLAQDD452 pKa = 3.83IAAGHH457 pKa = 6.21AVNNPSVSVSFPTDD471 pKa = 2.78NSTSSQITRR480 pKa = 11.84IQSSLVALQNLNGPGQGCPAASTTLSAQLAALQGG514 pKa = 3.57
Molecular weight: 50.91 kDa
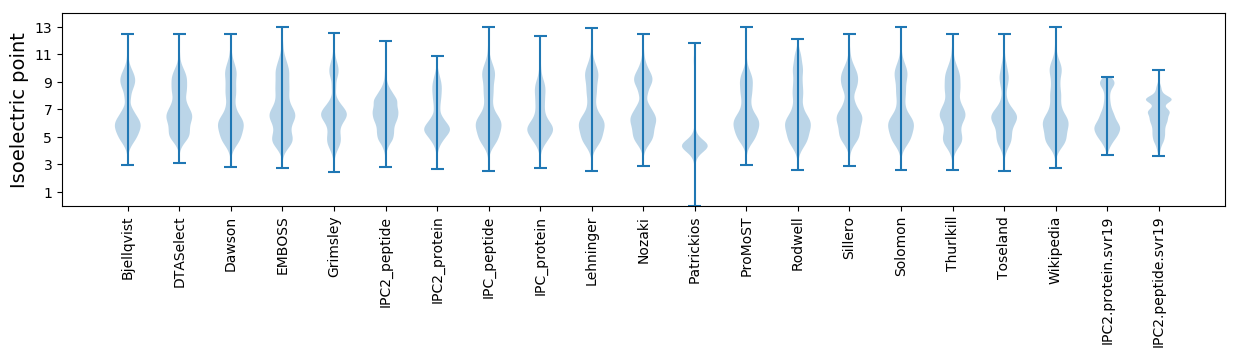
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S4KXC8|A0A4S4KXC8_9AGAM Uncharacterized protein OS=Phellinidium pouzarii OX=167371 GN=EW145_g6376 PE=4 SV=1
MM1 pKa = 7.44SAPAAANTRR10 pKa = 11.84RR11 pKa = 11.84WAPTSDD17 pKa = 3.6RR18 pKa = 11.84PNSIDD23 pKa = 3.47PAPVHH28 pKa = 6.14SRR30 pKa = 11.84PAFSSFSSFSSSISSSLSSAFGVGSSRR57 pKa = 11.84HH58 pKa = 5.27SRR60 pKa = 11.84SNSNATQSSQLNPNSHH76 pKa = 7.23PLRR79 pKa = 11.84PASAAPTAHH88 pKa = 5.99GHH90 pKa = 6.35AQAKK94 pKa = 7.28WLPRR98 pKa = 11.84TLTPGPGXRR107 pKa = 11.84RR108 pKa = 11.84LLITHH113 pKa = 7.12APAIVRR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84ARR123 pKa = 11.84LPPPQPQQLTRR134 pKa = 11.84LQTTXPLPPLPPLPXSRR151 pKa = 11.84QHH153 pKa = 6.52SSVLVGYY160 pKa = 7.03TKK162 pKa = 10.48PRR164 pKa = 11.84AKK166 pKa = 10.51GG167 pKa = 3.21
MM1 pKa = 7.44SAPAAANTRR10 pKa = 11.84RR11 pKa = 11.84WAPTSDD17 pKa = 3.6RR18 pKa = 11.84PNSIDD23 pKa = 3.47PAPVHH28 pKa = 6.14SRR30 pKa = 11.84PAFSSFSSFSSSISSSLSSAFGVGSSRR57 pKa = 11.84HH58 pKa = 5.27SRR60 pKa = 11.84SNSNATQSSQLNPNSHH76 pKa = 7.23PLRR79 pKa = 11.84PASAAPTAHH88 pKa = 5.99GHH90 pKa = 6.35AQAKK94 pKa = 7.28WLPRR98 pKa = 11.84TLTPGPGXRR107 pKa = 11.84RR108 pKa = 11.84LLITHH113 pKa = 7.12APAIVRR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84ARR123 pKa = 11.84LPPPQPQQLTRR134 pKa = 11.84LQTTXPLPPLPPLPXSRR151 pKa = 11.84QHH153 pKa = 6.52SSVLVGYY160 pKa = 7.03TKK162 pKa = 10.48PRR164 pKa = 11.84AKK166 pKa = 10.51GG167 pKa = 3.21
Molecular weight: 17.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4555891 |
33 |
4853 |
523.8 |
57.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.591 ± 0.023 | 1.174 ± 0.008 |
5.744 ± 0.016 | 6.032 ± 0.025 |
3.823 ± 0.014 | 6.226 ± 0.023 |
2.524 ± 0.011 | 4.927 ± 0.019 |
4.666 ± 0.022 | 9.183 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.064 ± 0.007 | 3.676 ± 0.013 |
6.198 ± 0.028 | 3.647 ± 0.018 |
6.197 ± 0.02 | 9.157 ± 0.038 |
6.05 ± 0.017 | 6.2 ± 0.019 |
1.276 ± 0.009 | 2.599 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
