
Oceanibacterium hippocampi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sneathiellales; Sneathiellaceae; Oceanibacterium
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
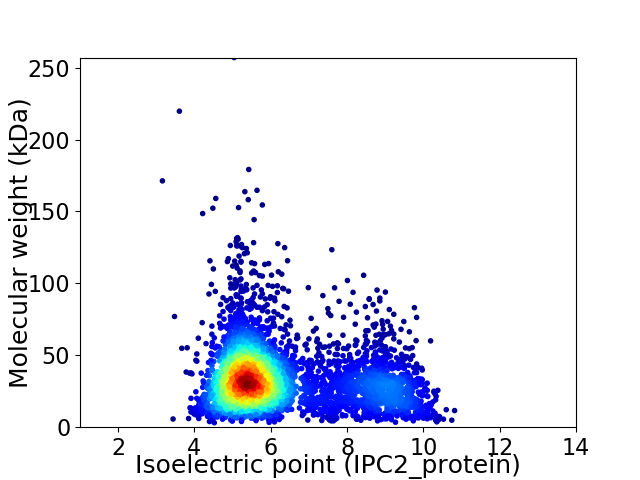
Virtual 2D-PAGE plot for 4524 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y5TB39|A0A1Y5TB39_9PROT HTH-type transcriptional repressor ComR OS=Oceanibacterium hippocampi OX=745714 GN=comR_2 PE=4 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84RR3 pKa = 11.84LLFGTTALGAVAFAAATAQAAEE25 pKa = 4.81PIQIGVGGFFSAFGVYY41 pKa = 10.11VNEE44 pKa = 4.91DD45 pKa = 3.64DD46 pKa = 4.95GAGEE50 pKa = 4.22PGAGTRR56 pKa = 11.84DD57 pKa = 3.61YY58 pKa = 11.15YY59 pKa = 10.85LARR62 pKa = 11.84EE63 pKa = 4.25GEE65 pKa = 4.47IIFSGSTVLDD75 pKa = 3.29NGLRR79 pKa = 11.84VGVNVQLEE87 pKa = 4.37AEE89 pKa = 4.39TCGDD93 pKa = 4.07QIDD96 pKa = 3.96EE97 pKa = 4.82SYY99 pKa = 11.8VFLAGNWGEE108 pKa = 4.05IRR110 pKa = 11.84LGSDD114 pKa = 4.53DD115 pKa = 4.37PVTDD119 pKa = 3.97AFSTGYY125 pKa = 7.37MTVVPEE131 pKa = 4.6HH132 pKa = 6.89GINDD136 pKa = 3.54PTHH139 pKa = 5.58FHH141 pKa = 4.96VHH143 pKa = 5.89IGANSLEE150 pKa = 4.35TPITTLGIGGDD161 pKa = 4.07SEE163 pKa = 4.42KK164 pKa = 10.85VSYY167 pKa = 10.3FSPTFSGFQFGVSYY181 pKa = 10.44TPDD184 pKa = 3.09NCEE187 pKa = 3.73EE188 pKa = 4.44GGCGGTYY195 pKa = 10.13SGSEE199 pKa = 3.97TDD201 pKa = 4.2NDD203 pKa = 3.74PSEE206 pKa = 4.11QGDD209 pKa = 4.13IIEE212 pKa = 4.21VAAMYY217 pKa = 10.08EE218 pKa = 4.37GEE220 pKa = 4.01FRR222 pKa = 11.84GIGIALSTGYY232 pKa = 10.6SRR234 pKa = 11.84GEE236 pKa = 4.0LEE238 pKa = 4.65ADD240 pKa = 3.44AGGDD244 pKa = 3.47DD245 pKa = 4.41DD246 pKa = 5.05QEE248 pKa = 4.18VWGFGAVLSYY258 pKa = 10.39MDD260 pKa = 3.57WSVGGGFRR268 pKa = 11.84HH269 pKa = 7.35DD270 pKa = 4.32NLGSDD275 pKa = 3.19NFDD278 pKa = 3.27RR279 pKa = 11.84RR280 pKa = 11.84DD281 pKa = 3.02WTVGVNYY288 pKa = 10.49NPGPWGVSLEE298 pKa = 4.13YY299 pKa = 11.23GEE301 pKa = 5.67AEE303 pKa = 4.23QEE305 pKa = 4.08TAAGDD310 pKa = 3.77DD311 pKa = 3.62EE312 pKa = 4.68VRR314 pKa = 11.84GVEE317 pKa = 4.52LGFAYY322 pKa = 10.14AVGPGVTFGAALQYY336 pKa = 10.36WDD338 pKa = 5.05LEE340 pKa = 4.56SGANNPAAEE349 pKa = 4.03NEE351 pKa = 4.0AWVVSVGTMLEE362 pKa = 4.21FF363 pKa = 4.88
MM1 pKa = 7.63RR2 pKa = 11.84RR3 pKa = 11.84LLFGTTALGAVAFAAATAQAAEE25 pKa = 4.81PIQIGVGGFFSAFGVYY41 pKa = 10.11VNEE44 pKa = 4.91DD45 pKa = 3.64DD46 pKa = 4.95GAGEE50 pKa = 4.22PGAGTRR56 pKa = 11.84DD57 pKa = 3.61YY58 pKa = 11.15YY59 pKa = 10.85LARR62 pKa = 11.84EE63 pKa = 4.25GEE65 pKa = 4.47IIFSGSTVLDD75 pKa = 3.29NGLRR79 pKa = 11.84VGVNVQLEE87 pKa = 4.37AEE89 pKa = 4.39TCGDD93 pKa = 4.07QIDD96 pKa = 3.96EE97 pKa = 4.82SYY99 pKa = 11.8VFLAGNWGEE108 pKa = 4.05IRR110 pKa = 11.84LGSDD114 pKa = 4.53DD115 pKa = 4.37PVTDD119 pKa = 3.97AFSTGYY125 pKa = 7.37MTVVPEE131 pKa = 4.6HH132 pKa = 6.89GINDD136 pKa = 3.54PTHH139 pKa = 5.58FHH141 pKa = 4.96VHH143 pKa = 5.89IGANSLEE150 pKa = 4.35TPITTLGIGGDD161 pKa = 4.07SEE163 pKa = 4.42KK164 pKa = 10.85VSYY167 pKa = 10.3FSPTFSGFQFGVSYY181 pKa = 10.44TPDD184 pKa = 3.09NCEE187 pKa = 3.73EE188 pKa = 4.44GGCGGTYY195 pKa = 10.13SGSEE199 pKa = 3.97TDD201 pKa = 4.2NDD203 pKa = 3.74PSEE206 pKa = 4.11QGDD209 pKa = 4.13IIEE212 pKa = 4.21VAAMYY217 pKa = 10.08EE218 pKa = 4.37GEE220 pKa = 4.01FRR222 pKa = 11.84GIGIALSTGYY232 pKa = 10.6SRR234 pKa = 11.84GEE236 pKa = 4.0LEE238 pKa = 4.65ADD240 pKa = 3.44AGGDD244 pKa = 3.47DD245 pKa = 4.41DD246 pKa = 5.05QEE248 pKa = 4.18VWGFGAVLSYY258 pKa = 10.39MDD260 pKa = 3.57WSVGGGFRR268 pKa = 11.84HH269 pKa = 7.35DD270 pKa = 4.32NLGSDD275 pKa = 3.19NFDD278 pKa = 3.27RR279 pKa = 11.84RR280 pKa = 11.84DD281 pKa = 3.02WTVGVNYY288 pKa = 10.49NPGPWGVSLEE298 pKa = 4.13YY299 pKa = 11.23GEE301 pKa = 5.67AEE303 pKa = 4.23QEE305 pKa = 4.08TAAGDD310 pKa = 3.77DD311 pKa = 3.62EE312 pKa = 4.68VRR314 pKa = 11.84GVEE317 pKa = 4.52LGFAYY322 pKa = 10.14AVGPGVTFGAALQYY336 pKa = 10.36WDD338 pKa = 5.05LEE340 pKa = 4.56SGANNPAAEE349 pKa = 4.03NEE351 pKa = 4.0AWVVSVGTMLEE362 pKa = 4.21FF363 pKa = 4.88
Molecular weight: 38.35 kDa
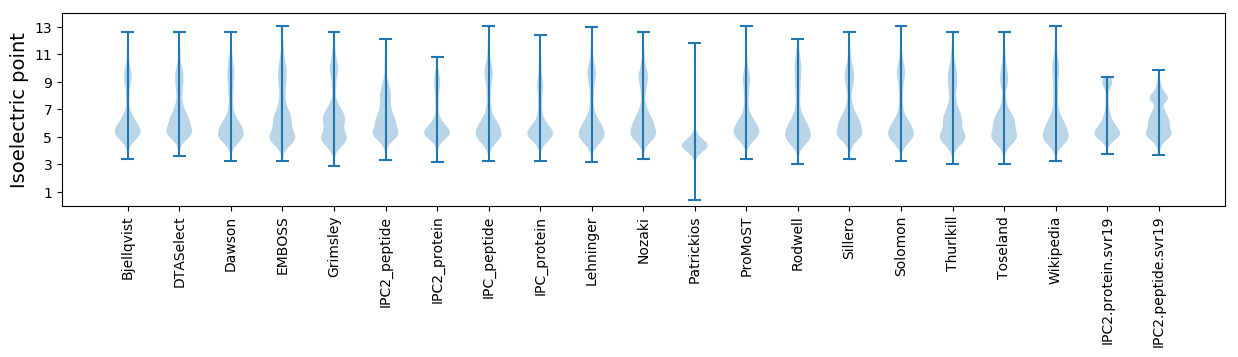
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y5U4H4|A0A1Y5U4H4_9PROT Holo-[acyl-carrier-protein] synthase OS=Oceanibacterium hippocampi OX=745714 GN=acpS PE=3 SV=1
MM1 pKa = 8.16ADD3 pKa = 3.4RR4 pKa = 11.84QLRR7 pKa = 11.84RR8 pKa = 11.84LDD10 pKa = 3.44RR11 pKa = 11.84QFRR14 pKa = 11.84ALSRR18 pKa = 11.84LAPGLAAAIGALRR31 pKa = 11.84GRR33 pKa = 11.84SGFLLRR39 pKa = 11.84FPLAVLLVIGGLLSFLPILGLWMVPLGLLLLAIDD73 pKa = 4.74LPVLRR78 pKa = 11.84PAVSAAIIRR87 pKa = 11.84LRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84WTRR94 pKa = 11.84WRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84SSS102 pKa = 2.83
MM1 pKa = 8.16ADD3 pKa = 3.4RR4 pKa = 11.84QLRR7 pKa = 11.84RR8 pKa = 11.84LDD10 pKa = 3.44RR11 pKa = 11.84QFRR14 pKa = 11.84ALSRR18 pKa = 11.84LAPGLAAAIGALRR31 pKa = 11.84GRR33 pKa = 11.84SGFLLRR39 pKa = 11.84FPLAVLLVIGGLLSFLPILGLWMVPLGLLLLAIDD73 pKa = 4.74LPVLRR78 pKa = 11.84PAVSAAIIRR87 pKa = 11.84LRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84WTRR94 pKa = 11.84WRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84SSS102 pKa = 2.83
Molecular weight: 11.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1462766 |
29 |
2388 |
323.3 |
35.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.916 ± 0.048 | 0.878 ± 0.013 |
5.952 ± 0.032 | 5.927 ± 0.031 |
3.699 ± 0.025 | 9.09 ± 0.032 |
1.998 ± 0.019 | 5.198 ± 0.027 |
2.686 ± 0.025 | 10.287 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.435 ± 0.016 | 2.428 ± 0.017 |
5.193 ± 0.024 | 2.642 ± 0.017 |
7.765 ± 0.041 | 4.957 ± 0.023 |
4.926 ± 0.025 | 7.463 ± 0.028 |
1.301 ± 0.013 | 2.261 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
